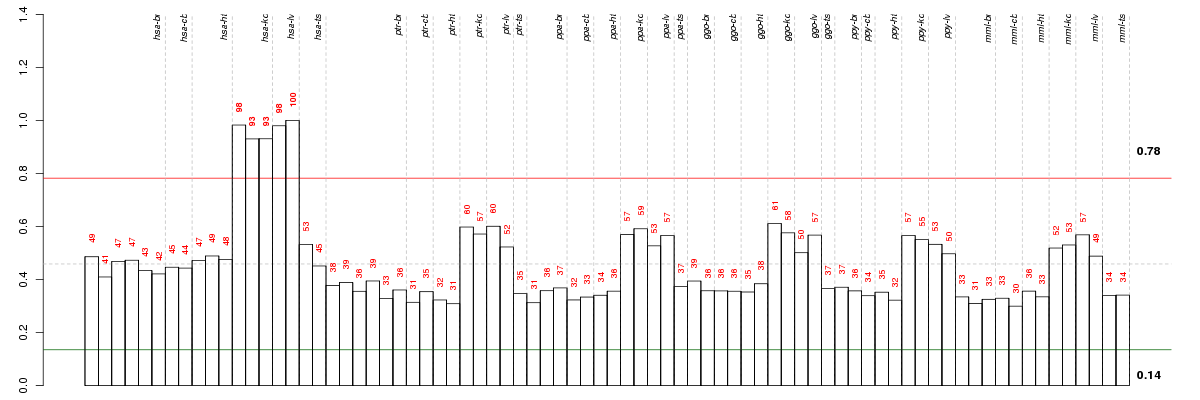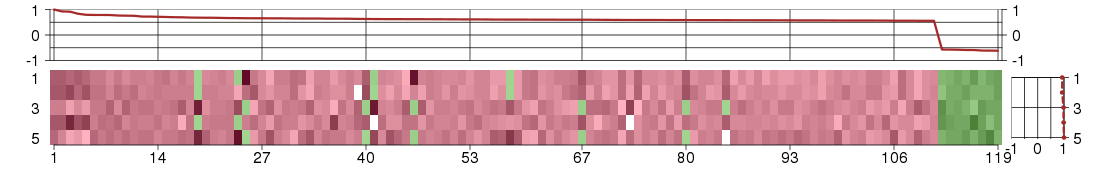

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

ANXA9annexin A9 (ENSG00000143412), score: 0.57 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: 0.59 APEX1APEX nuclease (multifunctional DNA repair enzyme) 1 (ENSG00000100823), score: 0.58 AQP3aquaporin 3 (Gill blood group) (ENSG00000165272), score: 0.64 ARSEarylsulfatase E (chondrodysplasia punctata 1) (ENSG00000157399), score: 0.72 ASPGasparaginase homolog (S. cerevisiae) (ENSG00000166183), score: 0.58 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: 0.6 B3GNT3UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 (ENSG00000179913), score: 0.79 C14orf21chromosome 14 open reading frame 21 (ENSG00000196943), score: 0.56 C14orf45chromosome 14 open reading frame 45 (ENSG00000119636), score: 0.65 C1orf186chromosome 1 open reading frame 186 (ENSG00000196533), score: 0.92 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: 0.59 C5orf15chromosome 5 open reading frame 15 (ENSG00000113583), score: 0.6 CBLCCas-Br-M (murine) ecotropic retroviral transforming sequence c (ENSG00000142273), score: 0.58 CCNT2cyclin T2 (ENSG00000082258), score: -0.59 CDHR2cadherin-related family member 2 (ENSG00000074276), score: 0.71 CDHR5cadherin-related family member 5 (ENSG00000099834), score: 0.68 CECR5cat eye syndrome chromosome region, candidate 5 (ENSG00000069998), score: 0.58 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: 0.57 CISD2CDGSH iron sulfur domain 2 (ENSG00000145354), score: 0.57 CLN6ceroid-lipofuscinosis, neuronal 6, late infantile, variant (ENSG00000128973), score: 0.59 CLPTM1LCLPTM1-like (ENSG00000049656), score: 0.58 CLRN3clarin 3 (ENSG00000180745), score: 0.7 CNGA1cyclic nucleotide gated channel alpha 1 (ENSG00000198515), score: 0.59 COASYCoA synthase (ENSG00000068120), score: 0.58 COQ4coenzyme Q4 homolog (S. cerevisiae) (ENSG00000167113), score: 0.61 CRYAAcrystallin, alpha A (ENSG00000160202), score: 0.65 CRYBB3crystallin, beta B3 (ENSG00000100053), score: 0.64 CWH43cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) (ENSG00000109182), score: 0.56 DHRS2dehydrogenase/reductase (SDR family) member 2 (ENSG00000100867), score: 0.62 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.56 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (ENSG00000160305), score: -0.57 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.72 DUSP23dual specificity phosphatase 23 (ENSG00000158716), score: 0.57 EEF1Geukaryotic translation elongation factor 1 gamma (ENSG00000149016), score: 0.67 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.75 ERGIC1endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 (ENSG00000113719), score: 0.6 FAM151Afamily with sequence similarity 151, member A (ENSG00000162391), score: 0.6 FBXO17F-box protein 17 (ENSG00000104835), score: 0.68 FTCDformiminotransferase cyclodeaminase (ENSG00000160282), score: 0.64 GAL3ST2galactose-3-O-sulfotransferase 2 (ENSG00000154252), score: 0.6 GANABglucosidase, alpha; neutral AB (ENSG00000089597), score: 0.59 GGT6gamma-glutamyltransferase 6 (ENSG00000167741), score: 0.58 GLTPD2glycolipid transfer protein domain containing 2 (ENSG00000182327), score: 0.66 GP1BAglycoprotein Ib (platelet), alpha polypeptide (ENSG00000185245), score: 0.6 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 1 HDAC6histone deacetylase 6 (ENSG00000094631), score: 0.61 HDHD3haloacid dehalogenase-like hydrolase domain containing 3 (ENSG00000119431), score: 0.61 HEATR6HEAT repeat containing 6 (ENSG00000068097), score: -0.61 IL17RBinterleukin 17 receptor B (ENSG00000056736), score: 0.65 IL17REinterleukin 17 receptor E (ENSG00000163701), score: 0.61 IL1RL2interleukin 1 receptor-like 2 (ENSG00000115598), score: 0.58 IL22RA1interleukin 22 receptor, alpha 1 (ENSG00000142677), score: 0.65 IL9Rinterleukin 9 receptor (ENSG00000124334), score: 0.6 KCNE3potassium voltage-gated channel, Isk-related family, member 3 (ENSG00000175538), score: 0.59 KCNH6potassium voltage-gated channel, subfamily H (eag-related), member 6 (ENSG00000173826), score: 0.6 LASS2LAG1 homolog, ceramide synthase 2 (ENSG00000143418), score: 0.57 LYPD2LY6/PLAUR domain containing 2 (ENSG00000197353), score: 0.66 MIFmacrophage migration inhibitory factor (glycosylation-inhibiting factor) (ENSG00000133460), score: 0.6 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.6 NCCRP1non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) (ENSG00000188505), score: 0.58 NPBneuropeptide B (ENSG00000183979), score: 0.62 OXER1oxoeicosanoid (OXE) receptor 1 (ENSG00000162881), score: 0.57 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.71 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.63 PEPDpeptidase D (ENSG00000124299), score: 0.6 PGAM5phosphoglycerate mutase family member 5 (ENSG00000176894), score: 0.65 PKLRpyruvate kinase, liver and RBC (ENSG00000143627), score: 0.62 PLEK2pleckstrin 2 (ENSG00000100558), score: 0.59 PLEKHJ1pleckstrin homology domain containing, family J member 1 (ENSG00000104886), score: 0.62 POFUT1protein O-fucosyltransferase 1 (ENSG00000101346), score: 0.61 PRRG4proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) (ENSG00000135378), score: 0.61 PRSS8protease, serine, 8 (ENSG00000052344), score: 0.65 PSMD9proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 (ENSG00000110801), score: 0.57 RAB17RAB17, member RAS oncogene family (ENSG00000124839), score: 0.61 RAB8ARAB8A, member RAS oncogene family (ENSG00000167461), score: 0.56 RALGAPA2Ral GTPase activating protein, alpha subunit 2 (catalytic) (ENSG00000188559), score: 0.58 RBP2retinol binding protein 2, cellular (ENSG00000114113), score: 0.62 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: 0.62 SESTD1SEC14 and spectrin domains 1 (ENSG00000187231), score: -0.61 SHHsonic hedgehog (ENSG00000164690), score: 0.65 SKAP1src kinase associated phosphoprotein 1 (ENSG00000141293), score: 0.58 SLA2Src-like-adaptor 2 (ENSG00000101082), score: 0.62 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (ENSG00000088386), score: 0.79 SLC16A13solute carrier family 16, member 13 (monocarboxylic acid transporter 13) (ENSG00000174327), score: 0.62 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.69 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.78 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.59 SLC25A42solute carrier family 25, member 42 (ENSG00000181035), score: 0.64 SLC39A5solute carrier family 39 (metal ion transporter), member 5 (ENSG00000139540), score: 0.79 SLC47A1solute carrier family 47, member 1 (ENSG00000142494), score: 0.57 SNX22sorting nexin 22 (ENSG00000157734), score: 0.62 ST13suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) (ENSG00000100380), score: 0.66 TCF4transcription factor 4 (ENSG00000196628), score: -0.57 TH1LTH1-like (Drosophila) (ENSG00000101158), score: 0.76 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.58 TM7SF3transmembrane 7 superfamily member 3 (ENSG00000064115), score: 0.58 TMED4transmembrane emp24 protein transport domain containing 4 (ENSG00000158604), score: 0.6 TMEM115transmembrane protein 115 (ENSG00000126062), score: 0.56 TMEM150Btransmembrane protein 150B (ENSG00000180061), score: 0.66 TMEM177transmembrane protein 177 (ENSG00000144120), score: 0.57 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.76 TRAP1TNF receptor-associated protein 1 (ENSG00000126602), score: 0.58 TRIM10tripartite motif-containing 10 (ENSG00000204613), score: 0.83 TRIM15tripartite motif-containing 15 (ENSG00000204610), score: 0.92 TRIP6thyroid hormone receptor interactor 6 (ENSG00000087077), score: 0.68 TRPC7transient receptor potential cation channel, subfamily C, member 7 (ENSG00000069018), score: 0.66 TSPAN33tetraspanin 33 (ENSG00000158457), score: 0.57 TSPO2translocator protein 2 (ENSG00000112212), score: 0.59 TUBAL3tubulin, alpha-like 3 (ENSG00000178462), score: 0.63 UGT2B7UDP glucuronosyltransferase 2 family, polypeptide B7 (ENSG00000171234), score: 0.56 USH2AUsher syndrome 2A (autosomal recessive, mild) (ENSG00000042781), score: 0.63 VAV2vav 2 guanine nucleotide exchange factor (ENSG00000160293), score: 0.57 ZC3H6zinc finger CCCH-type containing 6 (ENSG00000188177), score: -0.59 ZG16zymogen granule protein 16 homolog (rat) (ENSG00000174992), score: 0.68 ZGPATzinc finger, CCCH-type with G patch domain (ENSG00000197114), score: 0.59 ZNF518Bzinc finger protein 518B (ENSG00000178163), score: -0.62
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |