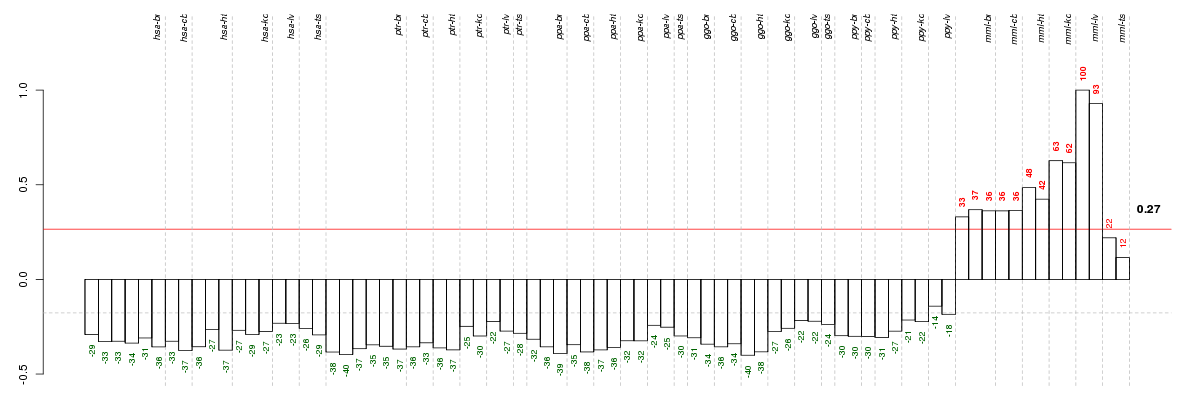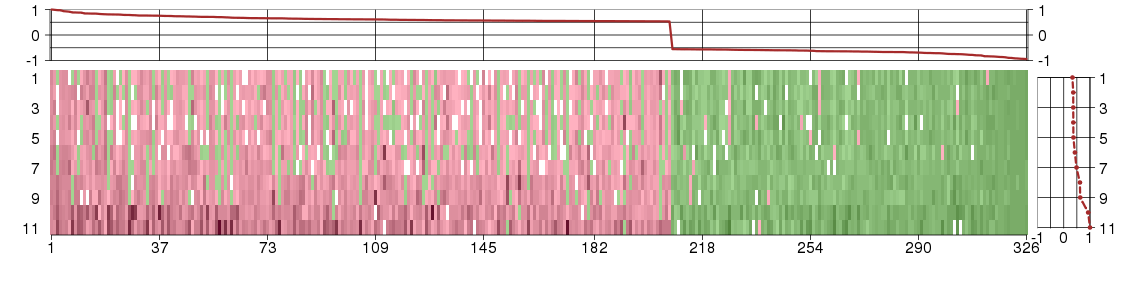

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
nucleoside phosphate metabolic process
The chemical reactions and pathways involving any phosphorylated nucleoside.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
nucleotide metabolic process
The chemical reactions and pathways involving a nucleotide, a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic nucleotides (nucleoside cyclic phosphates).
deoxyribonucleotide metabolic process
The chemical reactions and pathways involving a deoxyribonucleotide, a compound consisting of deoxyribonucleoside (a base linked to a deoxyribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05012 | 1.627e-02 | 1.539 | 7 | 72 | Parkinson's disease |
| 05010 | 2.979e-02 | 2.245 | 8 | 105 | Alzheimer's disease |
A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.56 ABCA4ATP-binding cassette, sub-family A (ABC1), member 4 (ENSG00000198691), score: 0.62 ABHD13abhydrolase domain containing 13 (ENSG00000139826), score: 0.54 ABHD3abhydrolase domain containing 3 (ENSG00000158201), score: 0.56 ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.84 ADAM12ADAM metallopeptidase domain 12 (ENSG00000148848), score: 0.88 AFPalpha-fetoprotein (ENSG00000081051), score: 0.76 AGR3anterior gradient homolog 3 (Xenopus laevis) (ENSG00000173467), score: 0.56 AIM1Labsent in melanoma 1-like (ENSG00000176092), score: 0.57 AIM2absent in melanoma 2 (ENSG00000163568), score: 0.68 AIREautoimmune regulator (ENSG00000160224), score: 0.72 AKR1CL1aldo-keto reductase family 1, member C-like 1 (ENSG00000196326), score: 0.79 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.6 ALOX12arachidonate 12-lipoxygenase (ENSG00000108839), score: 0.8 ALX3ALX homeobox 3 (ENSG00000156150), score: -0.58 ANO7anoctamin 7 (ENSG00000146205), score: 0.61 APAF1apoptotic peptidase activating factor 1 (ENSG00000120868), score: 0.55 APOA4apolipoprotein A-IV (ENSG00000110244), score: 0.75 AQP8aquaporin 8 (ENSG00000103375), score: 0.57 ARCN1archain 1 (ENSG00000095139), score: 0.71 ARIH2ariadne homolog 2 (Drosophila) (ENSG00000177479), score: -0.67 ARSBarylsulfatase B (ENSG00000113273), score: 0.59 ART4ADP-ribosyltransferase 4 (Dombrock blood group) (ENSG00000111339), score: 0.53 AS3MTarsenic (+3 oxidation state) methyltransferase (ENSG00000214435), score: 0.55 ATP8ATP synthase F0 subunit 8 (ENSG00000228253), score: -0.83 ATPBD4ATP binding domain 4 (ENSG00000134146), score: 0.57 AURKAIP1aurora kinase A interacting protein 1 (ENSG00000175756), score: -0.56 B3GAT2beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) (ENSG00000112309), score: -0.68 B3GNT7UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 (ENSG00000156966), score: 0.68 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (ENSG00000083123), score: 0.55 BIRC7baculoviral IAP repeat-containing 7 (ENSG00000101197), score: 0.83 C14orf126chromosome 14 open reading frame 126 (ENSG00000129480), score: 0.75 C14orf135chromosome 14 open reading frame 135 (ENSG00000126773), score: -0.57 C16orf91chromosome 16 open reading frame 91 (ENSG00000174109), score: 0.73 C17orf49chromosome 17 open reading frame 49 (ENSG00000161939), score: 0.76 C17orf90chromosome 17 open reading frame 90 (ENSG00000204237), score: -0.57 C19orf46chromosome 19 open reading frame 46 (ENSG00000181392), score: -0.66 C19orf53chromosome 19 open reading frame 53 (ENSG00000104979), score: -0.69 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: 0.54 C1orf177chromosome 1 open reading frame 177 (ENSG00000162398), score: 0.85 C1orf54chromosome 1 open reading frame 54 (ENSG00000118292), score: -0.56 C20orf29chromosome 20 open reading frame 29 (ENSG00000125843), score: 0.62 C20orf7chromosome 20 open reading frame 7 (ENSG00000101247), score: -0.78 C22orf15chromosome 22 open reading frame 15 (ENSG00000169314), score: 0.93 C22orf39chromosome 22 open reading frame 39 (ENSG00000242259), score: -0.75 C2orf43chromosome 2 open reading frame 43 (ENSG00000118961), score: 0.74 C2orf61chromosome 2 open reading frame 61 (ENSG00000239605), score: 0.63 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.59 C3orf26chromosome 3 open reading frame 26 (ENSG00000184220), score: -0.72 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.64 C4orf33chromosome 4 open reading frame 33 (ENSG00000151470), score: 0.57 C5orf45chromosome 5 open reading frame 45 (ENSG00000161010), score: -0.61 C6orf108chromosome 6 open reading frame 108 (ENSG00000112667), score: -0.74 C6orf52chromosome 6 open reading frame 52 (ENSG00000137434), score: 0.66 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: 0.62 C9orf102chromosome 9 open reading frame 102 (ENSG00000182150), score: 0.55 C9orf152chromosome 9 open reading frame 152 (ENSG00000188959), score: 0.88 C9orf6chromosome 9 open reading frame 6 (ENSG00000119328), score: 0.61 CABP4calcium binding protein 4 (ENSG00000175544), score: 0.63 CABP5calcium binding protein 5 (ENSG00000105507), score: 0.81 CACNA1Fcalcium channel, voltage-dependent, L type, alpha 1F subunit (ENSG00000102001), score: 0.58 CASP9caspase 9, apoptosis-related cysteine peptidase (ENSG00000132906), score: 0.59 CBARA1calcium binding atopy-related autoantigen 1 (ENSG00000107745), score: -0.67 CBY1chibby homolog 1 (Drosophila) (ENSG00000100211), score: -0.7 CCDC115coiled-coil domain containing 115 (ENSG00000136710), score: 0.6 CCDC137coiled-coil domain containing 137 (ENSG00000185298), score: -0.65 CCDC142coiled-coil domain containing 142 (ENSG00000135637), score: 0.82 CCDC42Bcoiled-coil domain containing 42B (ENSG00000186710), score: -0.64 CCL23chemokine (C-C motif) ligand 23 (ENSG00000167236), score: 0.64 CCL24chemokine (C-C motif) ligand 24 (ENSG00000106178), score: 0.61 CCL25chemokine (C-C motif) ligand 25 (ENSG00000131142), score: 0.88 CCNB1IP1cyclin B1 interacting protein 1 (ENSG00000100814), score: -0.64 CCR3chemokine (C-C motif) receptor 3 (ENSG00000183625), score: 0.55 CD1ECD1e molecule (ENSG00000158488), score: 0.73 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: -0.57 CDK5cyclin-dependent kinase 5 (ENSG00000164885), score: -0.8 CDKN2AIPNLCDKN2A interacting protein N-terminal like (ENSG00000237190), score: -0.87 CDV3CDV3 homolog (mouse) (ENSG00000091527), score: -0.9 CDX2caudal type homeobox 2 (ENSG00000165556), score: 0.55 CEACAM19carcinoembryonic antigen-related cell adhesion molecule 19 (ENSG00000186567), score: -0.64 CHCHD2coiled-coil-helix-coiled-coil-helix domain containing 2 (ENSG00000106153), score: -0.84 CHIC2cysteine-rich hydrophobic domain 2 (ENSG00000109220), score: 0.57 CHMP4Achromatin modifying protein 4A (ENSG00000100931), score: -0.64 CHRNA1cholinergic receptor, nicotinic, alpha 1 (muscle) (ENSG00000138435), score: 0.59 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.63 CLDN24claudin 24 (ENSG00000185758), score: 0.58 CLINT1clathrin interactor 1 (ENSG00000113282), score: 0.59 CNIHcornichon homolog (Drosophila) (ENSG00000100528), score: 0.63 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.63 CRYBA2crystallin, beta A2 (ENSG00000163499), score: 0.75 CXCR3chemokine (C-X-C motif) receptor 3 (ENSG00000186810), score: 0.55 CYP1A1cytochrome P450, family 1, subfamily A, polypeptide 1 (ENSG00000140465), score: 0.54 DEM1defects in morphology 1 homolog (S. cerevisiae) (ENSG00000164002), score: -0.59 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.69 DISP1dispatched homolog 1 (Drosophila) (ENSG00000154309), score: 0.55 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.7 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: -0.67 DSC3desmocollin 3 (ENSG00000134762), score: 0.78 DTYMKdeoxythymidylate kinase (thymidylate kinase) (ENSG00000168393), score: -0.65 EARS2glutamyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000103356), score: 0.54 EIF1ADeukaryotic translation initiation factor 1A domain containing (ENSG00000175376), score: 0.54 ELMOD3ELMO/CED-12 domain containing 3 (ENSG00000115459), score: 0.67 EME1essential meiotic endonuclease 1 homolog 1 (S. pombe) (ENSG00000154920), score: 0.66 ERHenhancer of rudimentary homolog (Drosophila) (ENSG00000100632), score: -0.62 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.81 FAM120AOSfamily with sequence similarity 120A opposite strand (ENSG00000188938), score: 0.73 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: 0.61 FAM173Bfamily with sequence similarity 173, member B (ENSG00000150756), score: -0.67 FAM184Bfamily with sequence similarity 184, member B (ENSG00000047662), score: 0.71 FAM189Bfamily with sequence similarity 189, member B (ENSG00000160767), score: -0.75 FAM36Afamily with sequence similarity 36, member A (ENSG00000203667), score: -0.56 FBXO22F-box protein 22 (ENSG00000167196), score: 0.54 FBXW8F-box and WD repeat domain containing 8 (ENSG00000174989), score: 0.62 FKTNfukutin (ENSG00000106692), score: 0.76 FLJ35220hypothetical protein FLJ35220 (ENSG00000173818), score: -0.65 FOXP3forkhead box P3 (ENSG00000049768), score: 0.71 FUNDC1FUN14 domain containing 1 (ENSG00000069509), score: -0.75 FXC1fracture callus 1 homolog (rat) (ENSG00000132286), score: 0.76 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.63 GGT5gamma-glutamyltransferase 5 (ENSG00000099998), score: -0.73 GINS1GINS complex subunit 1 (Psf1 homolog) (ENSG00000101003), score: 0.69 GLYATL3glycine-N-acyltransferase-like 3 (ENSG00000203972), score: 0.61 GNAT1guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 (ENSG00000114349), score: 0.59 GNAT2guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 (ENSG00000134183), score: 0.55 GON4Lgon-4-like (C. elegans) (ENSG00000116580), score: 0.54 GPC6glypican 6 (ENSG00000183098), score: 0.56 GPR113G protein-coupled receptor 113 (ENSG00000173567), score: 0.79 GPR152G protein-coupled receptor 152 (ENSG00000175514), score: 0.58 GPR172BG protein-coupled receptor 172B (ENSG00000132517), score: 0.6 GPR3G protein-coupled receptor 3 (ENSG00000181773), score: 0.54 GTF2F1general transcription factor IIF, polypeptide 1, 74kDa (ENSG00000125651), score: -0.92 HAS3hyaluronan synthase 3 (ENSG00000103044), score: 0.55 HAUS3HAUS augmin-like complex, subunit 3 (ENSG00000214367), score: -0.57 HDAC7histone deacetylase 7 (ENSG00000061273), score: -0.64 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.59 HEATR3HEAT repeat containing 3 (ENSG00000155393), score: 0.67 HEATR4HEAT repeat containing 4 (ENSG00000187105), score: -0.68 HFEhemochromatosis (ENSG00000010704), score: 0.62 HIVEP1human immunodeficiency virus type I enhancer binding protein 1 (ENSG00000095951), score: 0.59 HNRNPLheterogeneous nuclear ribonucleoprotein L (ENSG00000104824), score: -0.94 HRASLS2HRAS-like suppressor 2 (ENSG00000133328), score: 0.8 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (ENSG00000169087), score: -0.6 IFIT1Binterferon-induced protein with tetratricopeptide repeats 1B (ENSG00000204010), score: 0.99 IGDCC4immunoglobulin superfamily, DCC subclass, member 4 (ENSG00000103742), score: 0.66 IKBKEinhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon (ENSG00000143466), score: 0.55 INO80EINO80 complex subunit E (ENSG00000169592), score: -0.84 ITFG2integrin alpha FG-GAP repeat containing 2 (ENSG00000111203), score: 0.76 ITGAEintegrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) (ENSG00000083457), score: -0.67 IVDisovaleryl-CoA dehydrogenase (ENSG00000128928), score: 0.55 JMJD4jumonji domain containing 4 (ENSG00000081692), score: 0.55 KIAA0141KIAA0141 (ENSG00000081791), score: 0.57 KIAA1609KIAA1609 (ENSG00000140950), score: 0.71 KIAA1967KIAA1967 (ENSG00000158941), score: -0.59 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.62 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.57 KRT28keratin 28 (ENSG00000173908), score: 0.81 KRTAP10-8keratin associated protein 10-8 (ENSG00000187766), score: 0.62 LGMNlegumain (ENSG00000100600), score: 0.55 LIPMlipase, family member M (ENSG00000173239), score: 0.58 LMBR1limb region 1 homolog (mouse) (ENSG00000105983), score: -0.62 LOC100127905family with sequence similarity 165, member B pseudogene (ENSG00000198738), score: 1 LOC100287750hypothetical protein LOC100287750 (ENSG00000162585), score: -0.6 LOC100291552similar to DALR anticodon binding domain containing 3 (ENSG00000178149), score: -0.6 LUC7LLUC7-like (S. cerevisiae) (ENSG00000007392), score: -0.74 LYPD3LY6/PLAUR domain containing 3 (ENSG00000124466), score: 0.62 MAGEF1melanoma antigen family F, 1 (ENSG00000177383), score: -0.56 MAGIXMAGI family member, X-linked (ENSG00000017621), score: 0.55 MAKmale germ cell-associated kinase (ENSG00000111837), score: 0.67 MANSC1MANSC domain containing 1 (ENSG00000111261), score: 0.65 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.54 MAP3K7mitogen-activated protein kinase kinase kinase 7 (ENSG00000135341), score: -0.56 MAT2Bmethionine adenosyltransferase II, beta (ENSG00000038274), score: 0.58 MBD4methyl-CpG binding domain protein 4 (ENSG00000129071), score: -0.59 MCFD2multiple coagulation factor deficiency 2 (ENSG00000180398), score: 0.56 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.65 MCM3minichromosome maintenance complex component 3 (ENSG00000112118), score: -0.6 MCM7minichromosome maintenance complex component 7 (ENSG00000166508), score: -0.59 MEFVMediterranean fever (ENSG00000103313), score: 0.73 MLH1mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli) (ENSG00000076242), score: -0.56 MMP21matrix metallopeptidase 21 (ENSG00000154485), score: 0.58 MOBKL2AMOB1, Mps One Binder kinase activator-like 2A (yeast) (ENSG00000172081), score: -0.59 MPV17LMPV17 mitochondrial membrane protein-like (ENSG00000156968), score: 0.62 MRPL49mitochondrial ribosomal protein L49 (ENSG00000149792), score: 0.54 MS4A14membrane-spanning 4-domains, subfamily A, member 14 (ENSG00000166928), score: 0.61 MS4A8Bmembrane-spanning 4-domains, subfamily A, member 8B (ENSG00000166959), score: 0.61 MSLNmesothelin (ENSG00000102854), score: 0.54 MSLNLmesothelin-like (ENSG00000162006), score: 0.56 MST1Rmacrophage stimulating 1 receptor (c-met-related tyrosine kinase) (ENSG00000164078), score: 0.55 MT1Fmetallothionein 1F (ENSG00000198417), score: 0.54 MTCP1mature T-cell proliferation 1 (ENSG00000214827), score: -0.64 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.67 MUC6mucin 6, oligomeric mucus/gel-forming (ENSG00000184956), score: -0.6 NAA40N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) (ENSG00000110583), score: -0.85 NAT10N-acetyltransferase 10 (GCN5-related) (ENSG00000135372), score: -0.57 ND4NADH dehydrogenase, subunit 4 (complex I) (ENSG00000198886), score: -0.67 NDUFC2NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa (ENSG00000151366), score: -0.71 NDUFV3NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa (ENSG00000160194), score: -0.65 NET1neuroepithelial cell transforming 1 (ENSG00000173848), score: 0.54 NGDNneuroguidin, EIF4E binding protein (ENSG00000129460), score: -0.61 NGRNneugrin, neurite outgrowth associated (ENSG00000182768), score: -0.92 NME6non-metastatic cells 6, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000172113), score: 0.66 NR2C2APnuclear receptor 2C2-associated protein (ENSG00000184162), score: -0.65 NRF1nuclear respiratory factor 1 (ENSG00000106459), score: -0.57 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.64 NUDT15nudix (nucleoside diphosphate linked moiety X)-type motif 15 (ENSG00000136159), score: -0.83 NUMBnumb homolog (Drosophila) (ENSG00000133961), score: 0.55 NUPL2nucleoporin like 2 (ENSG00000136243), score: -0.71 OPTNoptineurin (ENSG00000123240), score: -0.64 OR10H3olfactory receptor, family 10, subfamily H, member 3 (ENSG00000171936), score: 0.69 OTUD1OTU domain containing 1 (ENSG00000165312), score: 0.57 OVGP1oviductal glycoprotein 1, 120kDa (ENSG00000085465), score: -0.56 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.64 PAEPprogestagen-associated endometrial protein (ENSG00000122133), score: 0.76 PAPLNpapilin, proteoglycan-like sulfated glycoprotein (ENSG00000100767), score: 0.56 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.59 PARP15poly (ADP-ribose) polymerase family, member 15 (ENSG00000173200), score: 0.62 PARS2prolyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000162396), score: 0.54 PCYOX1prenylcysteine oxidase 1 (ENSG00000116005), score: 0.55 PDCD2programmed cell death 2 (ENSG00000071994), score: 0.64 PDCD2Lprogrammed cell death 2-like (ENSG00000126249), score: -0.57 PDE12phosphodiesterase 12 (ENSG00000174840), score: 0.53 PIGHphosphatidylinositol glycan anchor biosynthesis, class H (ENSG00000100564), score: -0.58 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.61 PLA2G2Dphospholipase A2, group IID (ENSG00000117215), score: 0.84 PLEKHB2pleckstrin homology domain containing, family B (evectins) member 2 (ENSG00000115762), score: -0.8 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.59 POU1F1POU class 1 homeobox 1 (ENSG00000064835), score: 0.54 PPAPDC1Bphosphatidic acid phosphatase type 2 domain containing 1B (ENSG00000147535), score: 0.57 PRCCpapillary renal cell carcinoma (translocation-associated) (ENSG00000143294), score: -0.59 PRDM4PR domain containing 4 (ENSG00000110851), score: -0.6 PRDX1peroxiredoxin 1 (ENSG00000117450), score: -0.56 PRHOXNBparahox cluster neighbor (ENSG00000183463), score: 0.79 PRR4proline rich 4 (lacrimal) (ENSG00000111215), score: -0.65 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.88 PSKH2protein serine kinase H2 (ENSG00000147613), score: 0.71 PTCD2pentatricopeptide repeat domain 2 (ENSG00000049883), score: 0.65 PTGFRprostaglandin F receptor (FP) (ENSG00000122420), score: 0.63 QRSL1glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 (ENSG00000130348), score: 0.55 RAB1BRAB1B, member RAS oncogene family (ENSG00000174903), score: -0.94 RAB40ALRAB40A, member RAS oncogene family-like (ENSG00000102128), score: 0.56 RANBP10RAN binding protein 10 (ENSG00000141084), score: 0.54 RBM6RNA binding motif protein 6 (ENSG00000004534), score: -0.8 REC8REC8 homolog (yeast) (ENSG00000100918), score: -0.71 RGS13regulator of G-protein signaling 13 (ENSG00000127074), score: 0.7 RGS19regulator of G-protein signaling 19 (ENSG00000171700), score: 0.54 RHBDD3rhomboid domain containing 3 (ENSG00000100263), score: 0.56 RNASE1ribonuclease, RNase A family, 1 (pancreatic) (ENSG00000129538), score: -0.64 RPL36ALribosomal protein L36a-like (ENSG00000165502), score: -0.9 RPP40ribonuclease P/MRP 40kDa subunit (ENSG00000124787), score: 0.66 RTN4IP1reticulon 4 interacting protein 1 (ENSG00000130347), score: 0.54 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.58 SATL1spermidine/spermine N1-acetyl transferase-like 1 (ENSG00000184788), score: -0.78 SCLT1sodium channel and clathrin linker 1 (ENSG00000151466), score: 0.93 SCNM1sodium channel modifier 1 (ENSG00000163156), score: -0.67 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.64 SEC24ASEC24 family, member A (S. cerevisiae) (ENSG00000113615), score: 0.57 SEPT12septin 12 (ENSG00000140623), score: 0.71 SERINC4serine incorporator 4 (ENSG00000184716), score: 0.97 SERPINA12serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 (ENSG00000165953), score: 0.65 SHROOM3shroom family member 3 (ENSG00000138771), score: 0.56 SIP1survival of motor neuron protein interacting protein 1 (ENSG00000092208), score: -0.86 SIRT4sirtuin 4 (ENSG00000089163), score: 0.58 SLC16A11solute carrier family 16, member 11 (monocarboxylic acid transporter 11) (ENSG00000174326), score: 0.6 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (ENSG00000119899), score: 0.58 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.63 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (ENSG00000197496), score: 0.56 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.59 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.57 SNRPFsmall nuclear ribonucleoprotein polypeptide F (ENSG00000139343), score: -0.77 SNUPNsnurportin 1 (ENSG00000169371), score: -0.71 SPTLC1serine palmitoyltransferase, long chain base subunit 1 (ENSG00000090054), score: -0.56 SRD5A3steroid 5 alpha-reductase 3 (ENSG00000128039), score: 0.7 SRP14signal recognition particle 14kDa (homologous Alu RNA binding protein) (ENSG00000140319), score: -0.93 SRSF6serine/arginine-rich splicing factor 6 (ENSG00000124193), score: 0.65 STK16serine/threonine kinase 16 (ENSG00000115661), score: 0.57 STRA13stimulated by retinoic acid 13 homolog (mouse) (ENSG00000169689), score: -0.59 SYNCsyncoilin, intermediate filament protein (ENSG00000162520), score: -0.56 TAF11TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa (ENSG00000064995), score: -0.55 TAS2R14taste receptor, type 2, member 14 (ENSG00000212127), score: -0.66 TBX19T-box 19 (ENSG00000143178), score: 0.74 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.63 TCOF1Treacher Collins-Franceschetti syndrome 1 (ENSG00000070814), score: -0.59 TEP1telomerase-associated protein 1 (ENSG00000129566), score: 0.66 TK2thymidine kinase 2, mitochondrial (ENSG00000166548), score: -0.56 TLR6toll-like receptor 6 (ENSG00000174130), score: 0.77 TMEM104transmembrane protein 104 (ENSG00000109066), score: 0.66 TMEM167Btransmembrane protein 167B (ENSG00000215717), score: 0.57 TMEM19transmembrane protein 19 (ENSG00000139291), score: 0.62 TMEM201transmembrane protein 201 (ENSG00000188807), score: 0.57 TMEM61transmembrane protein 61 (ENSG00000143001), score: 0.56 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.61 TNNtenascin N (ENSG00000120332), score: 0.74 TRPM5transient receptor potential cation channel, subfamily M, member 5 (ENSG00000070985), score: 0.68 TSPAN11tetraspanin 11 (ENSG00000110900), score: 0.54 TTC21Btetratricopeptide repeat domain 21B (ENSG00000123607), score: -0.61 TTC24tetratricopeptide repeat domain 24 (ENSG00000187862), score: 0.84 TTC9Ctetratricopeptide repeat domain 9C (ENSG00000162222), score: -0.66 UBQLN1ubiquilin 1 (ENSG00000135018), score: 0.55 UBXN11UBX domain protein 11 (ENSG00000158062), score: -0.64 UCMAupper zone of growth plate and cartilage matrix associated (ENSG00000165623), score: 0.61 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.53 VILLvillin-like (ENSG00000136059), score: -0.58 VSIG10V-set and immunoglobulin domain containing 10 (ENSG00000176834), score: -0.68 WDR55WD repeat domain 55 (ENSG00000120314), score: 0.57 WDR73WD repeat domain 73 (ENSG00000177082), score: -0.6 WDSUB1WD repeat, sterile alpha motif and U-box domain containing 1 (ENSG00000196151), score: 0.54 XPCxeroderma pigmentosum, complementation group C (ENSG00000154767), score: 0.57 YAP1Yes-associated protein 1 (ENSG00000137693), score: -0.71 ZBED3zinc finger, BED-type containing 3 (ENSG00000132846), score: 0.57 ZBTB3zinc finger and BTB domain containing 3 (ENSG00000185670), score: 0.65 ZMYND19zinc finger, MYND-type containing 19 (ENSG00000165724), score: -0.61 ZNF114zinc finger protein 114 (ENSG00000178150), score: 0.84 ZNF146zinc finger protein 146 (ENSG00000167635), score: -0.86 ZNF16zinc finger protein 16 (ENSG00000170631), score: 0.8 ZNF226zinc finger protein 226 (ENSG00000167380), score: -0.69 ZNF408zinc finger protein 408 (ENSG00000175213), score: 0.54 ZNF496zinc finger protein 496 (ENSG00000162714), score: -0.6 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.76 ZSWIM3zinc finger, SWIM-type containing 3 (ENSG00000132801), score: 0.76
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_br_m2_ca1 | mml | br | m | 2 |
| mml_cb_m_ca1 | mml | cb | m | _ |
| mml_br_f_ca1 | mml | br | f | _ |
| mml_cb_f_ca1 | mml | cb | f | _ |
| mml_br_m1_ca1 | mml | br | m | 1 |
| mml_ht_f_ca1 | mml | ht | f | _ |
| mml_ht_m_ca1 | mml | ht | m | _ |
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |