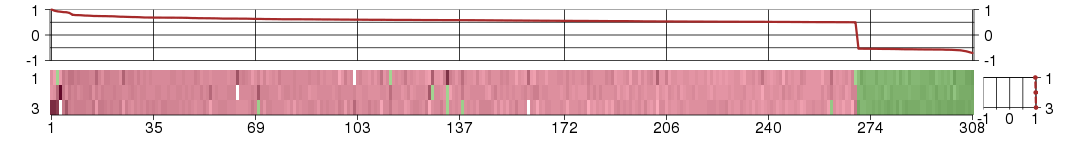

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of gene expression
Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The cellular synthesis of RNA on a template of DNA.
regulation of transcription
Any process that modulates the frequency, rate or extent of the synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
regulation of RNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving RNA.
transcription
The cellular synthesis of either RNA on a template of DNA or DNA on a template of RNA.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.
regulation of transcription, DNA-dependent
Any process that modulates the frequency, rate or extent of cellular DNA-dependent transcription.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
sequence-specific DNA binding transcription factor activity
Interacting selectively and non-covalently with a specific DNA sequence in order to modulate transcription. The transcription factor may or may not also interact selectively with a protein or macromolecular complex.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
sequence-specific DNA binding
Interacting selectively and non-covalently with DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding.
all
NA
AASDHaminoadipate-semialdehyde dehydrogenase (ENSG00000157426), score: 0.57 ACVR2Bactivin A receptor, type IIB (ENSG00000114739), score: 0.51 ADAM10ADAM metallopeptidase domain 10 (ENSG00000137845), score: 0.6 AFF4AF4/FMR2 family, member 4 (ENSG00000072364), score: 0.51 AMIGO3adhesion molecule with Ig-like domain 3 (ENSG00000176020), score: 0.76 ANKLE1ankyrin repeat and LEM domain containing 1 (ENSG00000160117), score: 0.54 ANKRD28ankyrin repeat domain 28 (ENSG00000206560), score: 0.5 ANXA11annexin A11 (ENSG00000122359), score: -0.56 ARSJarylsulfatase family, member J (ENSG00000180801), score: 0.61 ARVCFarmadillo repeat gene deleted in velocardiofacial syndrome (ENSG00000099889), score: 0.53 ATF7activating transcription factor 7 (ENSG00000170653), score: 0.72 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (ENSG00000112182), score: 0.57 BAHCC1BAH domain and coiled-coil containing 1 (ENSG00000171282), score: 0.52 BANK1B-cell scaffold protein with ankyrin repeats 1 (ENSG00000153064), score: 0.9 BARHL1BarH-like homeobox 1 (ENSG00000125492), score: 0.55 BARHL2BarH-like homeobox 2 (ENSG00000143032), score: 0.73 BCL2L10BCL2-like 10 (apoptosis facilitator) (ENSG00000137875), score: 0.52 BCL2L15BCL2-like 15 (ENSG00000188761), score: 0.59 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.6 BPTFbromodomain PHD finger transcription factor (ENSG00000171634), score: 0.59 BRWD3bromodomain and WD repeat domain containing 3 (ENSG00000165288), score: 0.57 BTN2A1butyrophilin, subfamily 2, member A1 (ENSG00000112763), score: 0.59 C14orf38chromosome 14 open reading frame 38 (ENSG00000151838), score: 0.52 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.62 C16orf11chromosome 16 open reading frame 11 (ENSG00000161992), score: 0.53 C20orf117chromosome 20 open reading frame 117 (ENSG00000149639), score: 0.58 C6orf115chromosome 6 open reading frame 115 (ENSG00000146386), score: -0.54 C6orf221chromosome 6 open reading frame 221 (ENSG00000203908), score: 0.59 C7orf42chromosome 7 open reading frame 42 (ENSG00000106609), score: -0.54 C8orf34chromosome 8 open reading frame 34 (ENSG00000165084), score: 0.75 CACNB4calcium channel, voltage-dependent, beta 4 subunit (ENSG00000182389), score: 0.51 CAPNS1calpain, small subunit 1 (ENSG00000126247), score: -0.57 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.68 CBLN3cerebellin 3 precursor (ENSG00000139899), score: 0.68 CCDC109Acoiled-coil domain containing 109A (ENSG00000156026), score: 0.55 CCNG2cyclin G2 (ENSG00000138764), score: 0.55 CCNJLcyclin J-like (ENSG00000135083), score: 0.61 CCR10chemokine (C-C motif) receptor 10 (ENSG00000184451), score: 0.51 CDAN1congenital dyserythropoietic anemia, type I (ENSG00000140326), score: 0.53 CDC25Bcell division cycle 25 homolog B (S. pombe) (ENSG00000101224), score: 0.51 CDC42BPGCDC42 binding protein kinase gamma (DMPK-like) (ENSG00000171219), score: 0.52 CDH15cadherin 15, type 1, M-cadherin (myotubule) (ENSG00000129910), score: 0.68 CDH24cadherin 24, type 2 (ENSG00000139880), score: 0.52 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.68 CDONCdon homolog (mouse) (ENSG00000064309), score: 0.62 CECR2cat eye syndrome chromosome region, candidate 2 (ENSG00000099954), score: 0.52 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: 0.61 CER1cerberus 1, cysteine knot superfamily, homolog (Xenopus laevis) (ENSG00000147869), score: 0.64 CERKLceramide kinase-like (ENSG00000188452), score: 0.6 CHCHD5coiled-coil-helix-coiled-coil-helix domain containing 5 (ENSG00000125611), score: -0.6 CHD7chromodomain helicase DNA binding protein 7 (ENSG00000171316), score: 0.65 CHN2chimerin (chimaerin) 2 (ENSG00000106069), score: 0.55 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.61 CIB1calcium and integrin binding 1 (calmyrin) (ENSG00000185043), score: -0.55 CLK4CDC-like kinase 4 (ENSG00000113240), score: 0.56 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: 0.58 CNPY1canopy 1 homolog (zebrafish) (ENSG00000146910), score: 0.57 CNTN6contactin 6 (ENSG00000134115), score: 0.62 COG2component of oligomeric golgi complex 2 (ENSG00000135775), score: 0.51 COL13A1collagen, type XIII, alpha 1 (ENSG00000197467), score: 0.76 COL19A1collagen, type XIX, alpha 1 (ENSG00000082293), score: 0.64 COL27A1collagen, type XXVII, alpha 1 (ENSG00000196739), score: 0.58 COL5A2collagen, type V, alpha 2 (ENSG00000204262), score: -0.54 CPLX4complexin 4 (ENSG00000166569), score: 0.93 CRLF3cytokine receptor-like factor 3 (ENSG00000176390), score: 0.77 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.74 CTCFCCCTC-binding factor (zinc finger protein) (ENSG00000102974), score: 0.59 CTNNB1catenin (cadherin-associated protein), beta 1, 88kDa (ENSG00000168036), score: 0.52 CXCR7chemokine (C-X-C motif) receptor 7 (ENSG00000144476), score: -0.57 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (ENSG00000165617), score: 0.53 DCAF16DDB1 and CUL4 associated factor 16 (ENSG00000163257), score: 0.52 DCLRE1BDNA cross-link repair 1B (ENSG00000118655), score: 0.64 DEFB136defensin, beta 136 (ENSG00000205884), score: 0.63 DGCR8DiGeorge syndrome critical region gene 8 (ENSG00000128191), score: 0.6 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: 0.6 DLG1discs, large homolog 1 (Drosophila) (ENSG00000075711), score: 0.5 DNMT3ADNA (cytosine-5-)-methyltransferase 3 alpha (ENSG00000119772), score: 0.54 ECE1endothelin converting enzyme 1 (ENSG00000117298), score: -0.71 EEDembryonic ectoderm development (ENSG00000074266), score: 0.55 EIF2C1eukaryotic translation initiation factor 2C, 1 (ENSG00000092847), score: 0.53 ELMO2engulfment and cell motility 2 (ENSG00000062598), score: 0.5 EN2engrailed homeobox 2 (ENSG00000164778), score: 0.71 EOMESeomesodermin (ENSG00000163508), score: 0.68 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: 0.59 ESPNLespin-like (ENSG00000144488), score: 0.58 ESYT3extended synaptotagmin-like protein 3 (ENSG00000158220), score: 0.61 ETV6ets variant 6 (ENSG00000139083), score: -0.57 EXOSC10exosome component 10 (ENSG00000171824), score: 0.61 EXPH5exophilin 5 (ENSG00000110723), score: 0.68 FAM110Bfamily with sequence similarity 110, member B (ENSG00000169122), score: 0.54 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: -0.55 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.69 FBXO38F-box protein 38 (ENSG00000145868), score: 0.5 FGF20fibroblast growth factor 20 (ENSG00000078579), score: 0.53 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.62 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.86 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.63 FKBP5FK506 binding protein 5 (ENSG00000096060), score: -0.54 FLT3fms-related tyrosine kinase 3 (ENSG00000122025), score: 0.57 FOXN2forkhead box N2 (ENSG00000170802), score: 0.56 FOXO1forkhead box O1 (ENSG00000150907), score: -0.54 FOXO3forkhead box O3 (ENSG00000118689), score: 0.58 FSTL5follistatin-like 5 (ENSG00000168843), score: 0.62 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.66 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.62 GALNT7UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (ENSG00000109586), score: 0.51 GCM2glial cells missing homolog 2 (Drosophila) (ENSG00000124827), score: 0.74 GDF7growth differentiation factor 7 (ENSG00000143869), score: 0.52 GDNFglial cell derived neurotrophic factor (ENSG00000168621), score: 0.58 GLCEglucuronic acid epimerase (ENSG00000138604), score: 0.64 GPATCH8G patch domain containing 8 (ENSG00000186566), score: 0.6 GPRIN3GPRIN family member 3 (ENSG00000185477), score: 0.63 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.58 GRID2IPglutamate receptor, ionotropic, delta 2 (Grid2) interacting protein (ENSG00000215045), score: 0.74 GRIN2Cglutamate receptor, ionotropic, N-methyl D-aspartate 2C (ENSG00000161509), score: 0.56 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.53 GRM4glutamate receptor, metabotropic 4 (ENSG00000124493), score: 0.62 HMG20Bhigh-mobility group 20B (ENSG00000064961), score: -0.56 IFFO2intermediate filament family orphan 2 (ENSG00000169991), score: 0.59 IL16interleukin 16 (lymphocyte chemoattractant factor) (ENSG00000172349), score: 0.54 IL23Ainterleukin 23, alpha subunit p19 (ENSG00000110944), score: 0.52 IMPG1interphotoreceptor matrix proteoglycan 1 (ENSG00000112706), score: 0.56 INCENPinner centromere protein antigens 135/155kDa (ENSG00000149503), score: 0.55 INO80INO80 homolog (S. cerevisiae) (ENSG00000128908), score: 0.62 IP6K2inositol hexakisphosphate kinase 2 (ENSG00000068745), score: 0.66 JMJD1Cjumonji domain containing 1C (ENSG00000171988), score: 0.68 KCNK10potassium channel, subfamily K, member 10 (ENSG00000100433), score: 0.57 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.53 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.7 KDM3Blysine (K)-specific demethylase 3B (ENSG00000120733), score: 0.53 KDM4Clysine (K)-specific demethylase 4C (ENSG00000107077), score: 0.64 KIAA0182KIAA0182 (ENSG00000131149), score: 0.64 KIAA0528KIAA0528 (ENSG00000111731), score: 0.53 KIAA0802KIAA0802 (ENSG00000168502), score: 0.55 KIAA0907KIAA0907 (ENSG00000132680), score: 0.51 KIAA1217KIAA1217 (ENSG00000120549), score: -0.58 KIFC3kinesin family member C3 (ENSG00000140859), score: -0.59 KLHL3kelch-like 3 (Drosophila) (ENSG00000146021), score: 0.51 KPRPkeratinocyte proline-rich protein (ENSG00000203786), score: 0.59 KRT39keratin 39 (ENSG00000196859), score: 1 KRT40keratin 40 (ENSG00000204889), score: 0.95 KRTAP22-2keratin associated protein 22-2 (ENSG00000206106), score: 0.58 LAMC2laminin, gamma 2 (ENSG00000058085), score: 0.59 LDB2LIM domain binding 2 (ENSG00000169744), score: -0.58 LMNB1lamin B1 (ENSG00000113368), score: 0.55 LOC146429putative solute carrier family 22 member ENSG00000182157 (ENSG00000182157), score: 0.58 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: 0.63 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.56 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.74 MAML1mastermind-like 1 (Drosophila) (ENSG00000161021), score: 0.68 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.69 MAMSTRMEF2 activating motif and SAP domain containing transcriptional regulator (ENSG00000176909), score: 0.6 MAN2B2mannosidase, alpha, class 2B, member 2 (ENSG00000013288), score: -0.54 MAXMYC associated factor X (ENSG00000125952), score: 0.58 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.68 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.54 MID1midline 1 (Opitz/BBB syndrome) (ENSG00000101871), score: 0.57 MLLmyeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) (ENSG00000118058), score: 0.56 MON2MON2 homolog (S. cerevisiae) (ENSG00000061987), score: 0.58 MPP3membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) (ENSG00000161647), score: 0.51 MPZmyelin protein zero (ENSG00000158887), score: 0.52 MRGPRDMAS-related GPR, member D (ENSG00000172938), score: 0.72 MRPL24mitochondrial ribosomal protein L24 (ENSG00000143314), score: -0.55 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.57 MRPS16mitochondrial ribosomal protein S16 (ENSG00000182180), score: -0.56 MSL1male-specific lethal 1 homolog (Drosophila) (ENSG00000188895), score: 0.51 MTBPMdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) binding protein, 104kDa (ENSG00000172167), score: 0.68 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.61 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (ENSG00000166886), score: 0.66 NHLH2nescient helix loop helix 2 (ENSG00000177551), score: 0.63 NHSL2NHS-like 2 (ENSG00000204131), score: 0.68 NIPAL2NIPA-like domain containing 2 (ENSG00000104361), score: 0.52 NKAIN1Na+/K+ transporting ATPase interacting 1 (ENSG00000084628), score: 0.64 NKTRnatural killer-tumor recognition sequence (ENSG00000114857), score: 0.53 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 0.79 NOTOnotochord homeobox (ENSG00000214513), score: 0.91 NPFFneuropeptide FF-amide peptide precursor (ENSG00000139574), score: 0.5 NRG4neuregulin 4 (ENSG00000169752), score: 0.55 NRIP2nuclear receptor interacting protein 2 (ENSG00000053702), score: 0.55 NYNRINNYN domain and retroviral integrase containing (ENSG00000205978), score: 0.54 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (ENSG00000107960), score: -0.59 ODZ1odz, odd Oz/ten-m homolog 1(Drosophila) (ENSG00000009694), score: 0.66 OGFRL1opioid growth factor receptor-like 1 (ENSG00000119900), score: 0.58 OTX2orthodenticle homeobox 2 (ENSG00000165588), score: 0.5 P4HA3prolyl 4-hydroxylase, alpha polypeptide III (ENSG00000149380), score: 0.58 PANK3pantothenate kinase 3 (ENSG00000120137), score: 0.53 PAPLiron/zinc purple acid phosphatase-like protein (ENSG00000183760), score: 0.57 PAX6paired box 6 (ENSG00000007372), score: 0.62 PAXIP1PAX interacting (with transcription-activation domain) protein 1 (ENSG00000157212), score: 0.61 PCBP4poly(rC) binding protein 4 (ENSG00000090097), score: 0.56 PDE8Aphosphodiesterase 8A (ENSG00000073417), score: -0.56 PHIPpleckstrin homology domain interacting protein (ENSG00000146247), score: 0.56 PISDphosphatidylserine decarboxylase (ENSG00000241878), score: 0.64 PKIBprotein kinase (cAMP-dependent, catalytic) inhibitor beta (ENSG00000135549), score: 0.72 PLA2G10phospholipase A2, group X (ENSG00000069764), score: 0.55 PLCB4phospholipase C, beta 4 (ENSG00000101333), score: 0.56 PLCXD3phosphatidylinositol-specific phospholipase C, X domain containing 3 (ENSG00000182836), score: 0.52 PLD5phospholipase D family, member 5 (ENSG00000180287), score: 0.54 PLK2polo-like kinase 2 (ENSG00000145632), score: -0.56 PLXND1plexin D1 (ENSG00000004399), score: -0.57 POLBpolymerase (DNA directed), beta (ENSG00000070501), score: 0.58 POLE4polymerase (DNA-directed), epsilon 4 (p12 subunit) (ENSG00000115350), score: -0.55 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (ENSG00000104356), score: 0.59 POP5processing of precursor 5, ribonuclease P/MRP subunit (S. cerevisiae) (ENSG00000167272), score: -0.54 POU2F2POU class 2 homeobox 2 (ENSG00000028277), score: 0.58 PPLperiplakin (ENSG00000118898), score: -0.55 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.63 PRPF3PRP3 pre-mRNA processing factor 3 homolog (S. cerevisiae) (ENSG00000117360), score: 0.52 PRPF38BPRP38 pre-mRNA processing factor 38 (yeast) domain containing B (ENSG00000134186), score: 0.55 PSMB7proteasome (prosome, macropain) subunit, beta type, 7 (ENSG00000136930), score: -0.57 PTCH1patched 1 (ENSG00000185920), score: 0.55 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.61 PTPN4protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) (ENSG00000088179), score: 0.51 PWWP2APWWP domain containing 2A (ENSG00000170234), score: 0.78 PYDC1PYD (pyrin domain) containing 1 (ENSG00000169900), score: 0.61 PYGO1pygopus homolog 1 (Drosophila) (ENSG00000171016), score: 0.59 QSOX2quiescin Q6 sulfhydryl oxidase 2 (ENSG00000165661), score: 0.62 RAB37RAB37, member RAS oncogene family (ENSG00000172794), score: 0.58 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.64 RELv-rel reticuloendotheliosis viral oncogene homolog (avian) (ENSG00000162924), score: 0.5 REV3LREV3-like, catalytic subunit of DNA polymerase zeta (yeast) (ENSG00000009413), score: 0.76 RFTN1raftlin, lipid raft linker 1 (ENSG00000131378), score: -0.64 RIPK2receptor-interacting serine-threonine kinase 2 (ENSG00000104312), score: -0.56 RIT2Ras-like without CAAX 2 (ENSG00000152214), score: 0.53 RMND5Arequired for meiotic nuclear division 5 homolog A (S. cerevisiae) (ENSG00000153561), score: 0.59 RNF146ring finger protein 146 (ENSG00000118518), score: 0.54 RTEL1regulator of telomere elongation helicase 1 (ENSG00000026036), score: 0.6 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: 0.51 SEC31BSEC31 homolog B (S. cerevisiae) (ENSG00000075826), score: 0.55 SEL1L3sel-1 suppressor of lin-12-like 3 (C. elegans) (ENSG00000091490), score: 0.5 SETBP1SET binding protein 1 (ENSG00000152217), score: 0.52 SETD5SET domain containing 5 (ENSG00000168137), score: 0.66 SF3B3splicing factor 3b, subunit 3, 130kDa (ENSG00000189091), score: 0.59 SFRS18splicing factor, arginine/serine-rich 18 (ENSG00000132424), score: 0.52 SGK223homolog of rat pragma of Rnd2 (ENSG00000182319), score: 0.53 SHKBP1SH3KBP1 binding protein 1 (ENSG00000160410), score: -0.54 SHPRHSNF2 histone linker PHD RING helicase (ENSG00000146414), score: 0.51 SIDT1SID1 transmembrane family, member 1 (ENSG00000072858), score: 0.52 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.51 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.6 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.69 SLC6A5solute carrier family 6 (neurotransmitter transporter, glycine), member 5 (ENSG00000165970), score: 0.59 SNRKSNF related kinase (ENSG00000163788), score: 0.71 SNX25sorting nexin 25 (ENSG00000109762), score: 0.59 SOCS5suppressor of cytokine signaling 5 (ENSG00000171150), score: 0.58 SP4Sp4 transcription factor (ENSG00000105866), score: 0.6 SPHKAPSPHK1 interactor, AKAP domain containing (ENSG00000153820), score: 0.52 SPIN3spindlin family, member 3 (ENSG00000204271), score: 0.51 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.6 SPRsepiapterin reductase (7,8-dihydrobiopterin:NADP+ oxidoreductase) (ENSG00000116096), score: -0.56 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.55 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.7 SURF1surfeit 1 (ENSG00000148290), score: -0.57 SYCP2Lsynaptonemal complex protein 2-like (ENSG00000153157), score: 0.75 SYNE1spectrin repeat containing, nuclear envelope 1 (ENSG00000131018), score: 0.53 SYT2synaptotagmin II (ENSG00000143858), score: 0.5 TAB1TGF-beta activated kinase 1/MAP3K7 binding protein 1 (ENSG00000100324), score: 0.56 TAS2R50taste receptor, type 2, member 50 (ENSG00000212126), score: 0.59 TEX264testis expressed 264 (ENSG00000164081), score: -0.55 TFAP2Etranscription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) (ENSG00000116819), score: 0.62 TGM1transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) (ENSG00000092295), score: 0.65 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.64 TK1thymidine kinase 1, soluble (ENSG00000167900), score: 0.5 TLL1tolloid-like 1 (ENSG00000038295), score: 0.73 TMC2transmembrane channel-like 2 (ENSG00000149488), score: 0.89 TMEM147transmembrane protein 147 (ENSG00000105677), score: -0.57 TNK1tyrosine kinase, non-receptor, 1 (ENSG00000174292), score: 0.52 TP73tumor protein p73 (ENSG00000078900), score: 0.6 TPRNtaperin (ENSG00000176058), score: 0.54 TRANK1tetratricopeptide repeat and ankyrin repeat containing 1 (ENSG00000168016), score: 0.6 TRIM11tripartite motif-containing 11 (ENSG00000154370), score: 0.78 TRIM17tripartite motif-containing 17 (ENSG00000162931), score: 0.59 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.68 TRMT6tRNA methyltransferase 6 homolog (S. cerevisiae) (ENSG00000089195), score: 0.54 TSEN34tRNA splicing endonuclease 34 homolog (S. cerevisiae) (ENSG00000170892), score: -0.58 UBASH3Bubiquitin associated and SH3 domain containing B (ENSG00000154127), score: 0.57 ULK3unc-51-like kinase 3 (C. elegans) (ENSG00000140474), score: 0.55 UNC13Bunc-13 homolog B (C. elegans) (ENSG00000198722), score: -0.67 UPF0639UPF0639 protein (ENSG00000175985), score: 0.6 USP48ubiquitin specific peptidase 48 (ENSG00000090686), score: 0.57 USTuronyl-2-sulfotransferase (ENSG00000111962), score: 0.65 VAT1Lvesicle amine transport protein 1 homolog (T. californica)-like (ENSG00000171724), score: 0.52 VPS45vacuolar protein sorting 45 homolog (S. cerevisiae) (ENSG00000136631), score: 0.51 VSX1visual system homeobox 1 (ENSG00000100987), score: 0.7 VWA5Avon Willebrand factor A domain containing 5A (ENSG00000110002), score: -0.62 WSCD2WSC domain containing 2 (ENSG00000075035), score: 0.52 XKR7XK, Kell blood group complex subunit-related family, member 7 (ENSG00000101321), score: 0.62 YTHDC1YTH domain containing 1 (ENSG00000083896), score: 0.57 ZFC3H1zinc finger, C3H1-type containing (ENSG00000133858), score: 0.53 ZIC2Zic family member 2 (odd-paired homolog, Drosophila) (ENSG00000043355), score: 0.6 ZIC4Zic family member 4 (ENSG00000174963), score: 0.69 ZIC5Zic family member 5 (odd-paired homolog, Drosophila) (ENSG00000139800), score: 0.67 ZNF132zinc finger protein 132 (ENSG00000131849), score: 0.51 ZNF141zinc finger protein 141 (ENSG00000131127), score: 0.54 ZNF157zinc finger protein 157 (ENSG00000147117), score: 0.56 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.52 ZNF296zinc finger protein 296 (ENSG00000170684), score: 0.53 ZNF337zinc finger protein 337 (ENSG00000130684), score: 0.66 ZNF397zinc finger protein 397 (ENSG00000186812), score: 0.61 ZNF45zinc finger protein 45 (ENSG00000124459), score: 0.58 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.61 ZNF671zinc finger protein 671 (ENSG00000083814), score: 0.53 ZNF772zinc finger protein 772 (ENSG00000197128), score: 0.56 ZNF774zinc finger protein 774 (ENSG00000196391), score: 0.56 ZNF776zinc finger protein 776 (ENSG00000152443), score: 0.52 ZZEF1zinc finger, ZZ-type with EF-hand domain 1 (ENSG00000074755), score: 0.51
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |