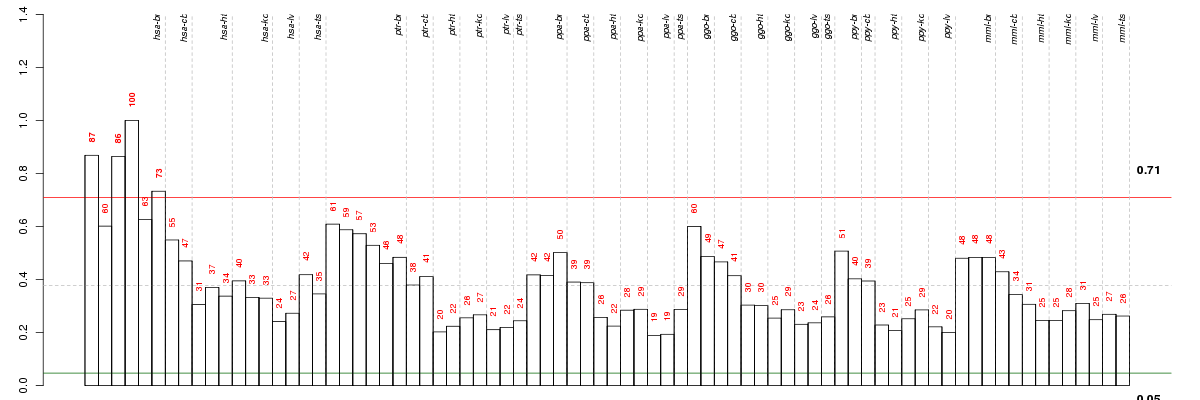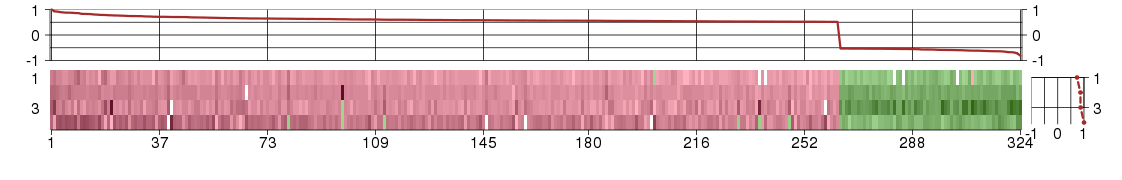

Under-expression is coded with green,
over-expression with red color.

regulation of action potential
Any process that modulates the frequency, rate or extent of action potential creation, propagation or termination. An action potential is a spike of membrane depolarization and repolarization that travels along the membrane of a cell.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
cellular ion homeostasis
Any process involved in the maintenance of an internal steady-state of ions at the level of a cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
ensheathment of neurons
The process whereby glial cells envelop neuronal cell bodies and/or axons to form an insulating layer. This can take the form of myelinating or non-myelinating ensheathment.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
axon ensheathment
Any process by which the axon of a neuron is insulated, and that insulation maintained, thereby preventing dispersion of the electrical signal.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
glial cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of a glial cell.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
regulation of action potential in neuron
The process that modulates the membrane potential involved in the propagation of a signal in a neuron.
cellular homeostasis
Any process involved in the maintenance of an internal steady-state at the level of the cell.
neurogenesis
Generation of cells within the nervous system.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
gliogenesis
The process by which glial cells are generated. This includes the production of glial progenitors and their differentiation into mature glia.
regulation of membrane potential
Any process that modulates the establishment or extent of a membrane potential, the electric potential existing across any membrane arising from charges in the membrane itself and from the charges present in the media on either side of the membrane.
homeostatic process
Any biological process involved in the maintenance of an internal steady-state.
ion homeostasis
Any process involved in the maintenance of an internal steady-state of ions within an organism or cell.
oligodendrocyte differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an oligodendrocyte. An oligodendrocyte is a type of glial cell involved in myelinating the axons of neurons in the central nervous system.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical.
cellular chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical at the level of the cell.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
all
NA
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
cellular homeostasis
Any process involved in the maintenance of an internal steady-state at the level of the cell.
ensheathment of neurons
The process whereby glial cells envelop neuronal cell bodies and/or axons to form an insulating layer. This can take the form of myelinating or non-myelinating ensheathment.
ensheathment of neurons
The process whereby glial cells envelop neuronal cell bodies and/or axons to form an insulating layer. This can take the form of myelinating or non-myelinating ensheathment.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
neurogenesis
Generation of cells within the nervous system.
oligodendrocyte differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an oligodendrocyte. An oligodendrocyte is a type of glial cell involved in myelinating the axons of neurons in the central nervous system.
cellular chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical at the level of the cell.
glial cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of a glial cell.
cellular ion homeostasis
Any process involved in the maintenance of an internal steady-state of ions at the level of a cell.
regulation of action potential in neuron
The process that modulates the membrane potential involved in the propagation of a signal in a neuron.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
myelin sheath
An electrically insulating fatty layer that surrounds the axons of many neurons. It is an outgrowth of glial cells: Schwann cells supply the myelin for peripheral neurons while oligodendrocytes supply it to those of the central nervous system.
compact myelin
The portion of the myelin sheath in which layers of cell membrane are tightly juxtaposed, completely excluding cytoplasm. The juxtaposed cytoplasmic surfaces form the major dense line, while the juxtaposed extracellular surfaces form the interperiod line visible in electron micrographs.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
compact myelin
The portion of the myelin sheath in which layers of cell membrane are tightly juxtaposed, completely excluding cytoplasm. The juxtaposed cytoplasmic surfaces form the major dense line, while the juxtaposed extracellular surfaces form the interperiod line visible in electron micrographs.

ABCA2ATP-binding cassette, sub-family A (ABC1), member 2 (ENSG00000107331), score: 0.63 ACOT8acyl-CoA thioesterase 8 (ENSG00000101473), score: 0.53 ADAMTS14ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ENSG00000138316), score: 0.54 ADAMTS4ADAM metallopeptidase with thrombospondin type 1 motif, 4 (ENSG00000158859), score: 0.58 ADARB2adenosine deaminase, RNA-specific, B2 (ENSG00000185736), score: 0.61 AGAP1ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 (ENSG00000157985), score: 0.52 AGPAT41-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) (ENSG00000026652), score: 0.56 AIDAaxin interactor, dorsalization associated (ENSG00000186063), score: -0.63 ANLNanillin, actin binding protein (ENSG00000011426), score: 0.55 APC2adenomatosis polyposis coli 2 (ENSG00000115266), score: 0.52 ARHGAP29Rho GTPase activating protein 29 (ENSG00000137962), score: -0.53 ARHGAP36Rho GTPase activating protein 36 (ENSG00000147256), score: 0.52 ARHGEF2Rho/Rac guanine nucleotide exchange factor (GEF) 2 (ENSG00000116584), score: 0.53 ASPRV1aspartic peptidase, retroviral-like 1 (ENSG00000244617), score: 0.56 ATL2atlastin GTPase 2 (ENSG00000119787), score: -0.64 ATP10BATPase, class V, type 10B (ENSG00000118322), score: 0.71 ATP11AATPase, class VI, type 11A (ENSG00000068650), score: 0.6 BACE1beta-site APP-cleaving enzyme 1 (ENSG00000186318), score: 0.58 BAIAP3BAI1-associated protein 3 (ENSG00000007516), score: 0.53 BAT5HLA-B associated transcript 5 (ENSG00000204422), score: 0.56 BIN1bridging integrator 1 (ENSG00000136717), score: 0.53 C11orf9chromosome 11 open reading frame 9 (ENSG00000124920), score: 0.68 C12orf34chromosome 12 open reading frame 34 (ENSG00000139438), score: 0.53 C16orf42chromosome 16 open reading frame 42 (ENSG00000007520), score: 0.53 C16orf70chromosome 16 open reading frame 70 (ENSG00000125149), score: 0.52 C1orf198chromosome 1 open reading frame 198 (ENSG00000119280), score: 0.71 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.59 C3orf16chromosome 3 open reading frame 16 (ENSG00000206199), score: 0.7 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.64 CAPN3calpain 3, (p94) (ENSG00000092529), score: 0.62 CAPN9calpain 9 (ENSG00000135773), score: 0.64 CCPG1cell cycle progression 1 (ENSG00000214882), score: -0.61 CCT8chaperonin containing TCP1, subunit 8 (theta) (ENSG00000156261), score: -0.82 CD22CD22 molecule (ENSG00000012124), score: 0.9 CD72CD72 molecule (ENSG00000137101), score: 0.52 CDC23cell division cycle 23 homolog (S. cerevisiae) (ENSG00000094880), score: -0.54 CDC26cell division cycle 26 homolog (S. cerevisiae) (ENSG00000176386), score: -0.63 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: 0.56 CDH20cadherin 20, type 2 (ENSG00000101542), score: 0.53 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.56 CDHR1cadherin-related family member 1 (ENSG00000148600), score: 0.63 CDK18cyclin-dependent kinase 18 (ENSG00000117266), score: 0.55 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 0.87 CHADLchondroadherin-like (ENSG00000100399), score: 0.8 CHST6carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 (ENSG00000183196), score: 0.59 CLCA4chloride channel accessory 4 (ENSG00000016602), score: 0.83 CLDN11claudin 11 (ENSG00000013297), score: 0.6 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.71 CLN8ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) (ENSG00000182372), score: 0.88 CLUL1clusterin-like 1 (retinal) (ENSG00000079101), score: 0.86 CMTM5CKLF-like MARVEL transmembrane domain containing 5 (ENSG00000166091), score: 0.6 CNGB1cyclic nucleotide gated channel beta 1 (ENSG00000070729), score: 0.53 CNP2',3'-cyclic nucleotide 3' phosphodiesterase (ENSG00000173786), score: 0.76 CNTN2contactin 2 (axonal) (ENSG00000184144), score: 0.56 COBLcordon-bleu homolog (mouse) (ENSG00000106078), score: 0.58 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.56 COL16A1collagen, type XVI, alpha 1 (ENSG00000084636), score: 0.52 COL9A2collagen, type IX, alpha 2 (ENSG00000049089), score: 0.57 CPNE2copine II (ENSG00000140848), score: 0.67 CRYBB1crystallin, beta B1 (ENSG00000100122), score: 0.71 CSRP1cysteine and glycine-rich protein 1 (ENSG00000159176), score: 0.56 CTNNAL1catenin (cadherin-associated protein), alpha-like 1 (ENSG00000119326), score: -0.58 CTR9Ctr9, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (ENSG00000198730), score: -0.53 CWC15CWC15 spliceosome-associated protein homolog (S. cerevisiae) (ENSG00000150316), score: -0.54 CWC22CWC22 spliceosome-associated protein homolog (S. cerevisiae) (ENSG00000163510), score: -0.55 CYB5D1cytochrome b5 domain containing 1 (ENSG00000182224), score: -0.73 DAAM2dishevelled associated activator of morphogenesis 2 (ENSG00000146122), score: 0.64 DEFB134defensin, beta 134 (ENSG00000205882), score: 0.78 DHPSdeoxyhypusine synthase (ENSG00000095059), score: 0.52 DIP2BDIP2 disco-interacting protein 2 homolog B (Drosophila) (ENSG00000066084), score: 0.54 DLL3delta-like 3 (Drosophila) (ENSG00000090932), score: 0.55 DMBT1deleted in malignant brain tumors 1 (ENSG00000187908), score: 0.79 DMRTA2DMRT-like family A2 (ENSG00000142700), score: 0.57 DNMBPdynamin binding protein (ENSG00000107554), score: -0.66 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.78 DRD4dopamine receptor D4 (ENSG00000069696), score: 0.52 DSCAML1Down syndrome cell adhesion molecule like 1 (ENSG00000177103), score: 0.55 DSELdermatan sulfate epimerase-like (ENSG00000171451), score: 0.65 DUSP11dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) (ENSG00000144048), score: -0.63 DYNC1I2dynein, cytoplasmic 1, intermediate chain 2 (ENSG00000077380), score: 0.54 EBAG9estrogen receptor binding site associated, antigen, 9 (ENSG00000147654), score: -0.54 EDIL3EGF-like repeats and discoidin I-like domains 3 (ENSG00000164176), score: 0.59 EFSembryonal Fyn-associated substrate (ENSG00000100842), score: 0.6 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.77 EML2echinoderm microtubule associated protein like 2 (ENSG00000125746), score: 0.63 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: 0.67 ERMNermin, ERM-like protein (ENSG00000136541), score: 0.55 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.65 FA2Hfatty acid 2-hydroxylase (ENSG00000103089), score: 0.68 FABP6fatty acid binding protein 6, ileal (ENSG00000170231), score: 0.74 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.64 FAM103A1family with sequence similarity 103, member A1 (ENSG00000169612), score: -0.64 FAM108B1family with sequence similarity 108, member B1 (ENSG00000107362), score: 0.65 FAM114A2family with sequence similarity 114, member A2 (ENSG00000055147), score: -0.55 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.82 FAM125Bfamily with sequence similarity 125, member B (ENSG00000196814), score: 0.71 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: -0.6 FAM53Bfamily with sequence similarity 53, member B (ENSG00000189319), score: 0.64 FAN1FANCD2/FANCI-associated nuclease 1 (ENSG00000198690), score: 0.56 FCHO1FCH domain only 1 (ENSG00000130475), score: 0.52 FGF22fibroblast growth factor 22 (ENSG00000070388), score: 0.62 FKBP15FK506 binding protein 15, 133kDa (ENSG00000119321), score: 0.52 FNBP1Lformin binding protein 1-like (ENSG00000137942), score: -0.6 FNTBfarnesyltransferase, CAAX box, beta (ENSG00000125954), score: 0.59 FOXRED2FAD-dependent oxidoreductase domain containing 2 (ENSG00000100350), score: 0.58 FSCN1fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) (ENSG00000075618), score: 0.54 GAB2GRB2-associated binding protein 2 (ENSG00000033327), score: 0.66 GABRQgamma-aminobutyric acid (GABA) receptor, theta (ENSG00000147402), score: 0.71 GAL3ST1galactose-3-O-sulfotransferase 1 (ENSG00000128242), score: 0.58 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (ENSG00000139629), score: 0.87 GALNTL2UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2 (ENSG00000131386), score: 0.53 GCAgrancalcin, EF-hand calcium binding protein (ENSG00000115271), score: 0.58 GIPC1GIPC PDZ domain containing family, member 1 (ENSG00000123159), score: 0.63 GIPRgastric inhibitory polypeptide receptor (ENSG00000010310), score: 0.71 GJD4gap junction protein, delta 4, 40.1kDa (ENSG00000177291), score: 0.55 GLDNgliomedin (ENSG00000186417), score: 0.63 GLRX3glutaredoxin 3 (ENSG00000108010), score: -0.53 GLTPglycolipid transfer protein (ENSG00000139433), score: 0.57 GNAI1guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 (ENSG00000127955), score: 0.54 GPAT2glycerol-3-phosphate acyltransferase 2, mitochondrial (ENSG00000186281), score: 0.57 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.61 GPR151G protein-coupled receptor 151 (ENSG00000173250), score: 0.53 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (ENSG00000170775), score: 0.63 GPR78G protein-coupled receptor 78 (ENSG00000155269), score: 0.77 GPRC5BG protein-coupled receptor, family C, group 5, member B (ENSG00000167191), score: 0.59 GREM1gremlin 1 (ENSG00000166923), score: 0.86 GRID1glutamate receptor, ionotropic, delta 1 (ENSG00000182771), score: 0.54 GTF2E2general transcription factor IIE, polypeptide 2, beta 34kDa (ENSG00000197265), score: -0.69 GUK1guanylate kinase 1 (ENSG00000143774), score: 0.66 HEATR1HEAT repeat containing 1 (ENSG00000119285), score: -0.59 HELQhelicase, POLQ-like (ENSG00000163312), score: -0.62 HHIPhedgehog interacting protein (ENSG00000164161), score: 0.53 HHIPL1HHIP-like 1 (ENSG00000182218), score: 0.66 HIPK2homeodomain interacting protein kinase 2 (ENSG00000064393), score: 0.53 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.69 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.74 IPO13importin 13 (ENSG00000117408), score: 0.57 ITGA2integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) (ENSG00000164171), score: 0.55 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: 0.6 JAK1Janus kinase 1 (ENSG00000162434), score: -0.55 JAM3junctional adhesion molecule 3 (ENSG00000166086), score: 0.56 KARSlysyl-tRNA synthetase (ENSG00000065427), score: -0.57 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.71 KELKell blood group, metallo-endopeptidase (ENSG00000197993), score: 0.52 KIAA1429KIAA1429 (ENSG00000164944), score: -0.54 KIAA1755KIAA1755 (ENSG00000149633), score: 0.63 KINKIN, antigenic determinant of recA protein homolog (mouse) (ENSG00000151657), score: -0.54 KIRREL2kin of IRRE like 2 (Drosophila) (ENSG00000126259), score: 0.65 KLHL32kelch-like 32 (Drosophila) (ENSG00000186231), score: 0.56 KLHL35kelch-like 35 (Drosophila) (ENSG00000149243), score: 0.59 KREMEN2kringle containing transmembrane protein 2 (ENSG00000131650), score: 1 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: 0.52 LAYNlayilin (ENSG00000204381), score: 0.58 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.83 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (ENSG00000107902), score: 0.74 LILRA4leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 (ENSG00000239961), score: 0.52 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.79 LOC100294412similar to KIAA0655 protein (ENSG00000130787), score: 0.56 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.93 LSM12LSM12 homolog (S. cerevisiae) (ENSG00000161654), score: -0.57 LYPD5LY6/PLAUR domain containing 5 (ENSG00000159871), score: 0.55 LZTS2leucine zipper, putative tumor suppressor 2 (ENSG00000107816), score: 0.52 MAGmyelin associated glycoprotein (ENSG00000105695), score: 0.62 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: -0.55 MALT1mucosa associated lymphoid tissue lymphoma translocation gene 1 (ENSG00000172175), score: -0.54 MAN2A2mannosidase, alpha, class 2A, member 2 (ENSG00000196547), score: 0.57 MARCH1membrane-associated ring finger (C3HC4) 1 (ENSG00000145416), score: 0.61 MARCKSL1MARCKS-like 1 (ENSG00000175130), score: 0.73 MED10mediator complex subunit 10 (ENSG00000133398), score: 0.55 MED20mediator complex subunit 20 (ENSG00000124641), score: -0.59 MEGF10multiple EGF-like-domains 10 (ENSG00000145794), score: 0.58 MGRN1mahogunin, ring finger 1 (ENSG00000102858), score: 0.54 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (ENSG00000165175), score: 0.6 MKRN3makorin ring finger protein 3 (ENSG00000179455), score: 0.63 MNAT1menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) (ENSG00000020426), score: -0.56 MOCS1molybdenum cofactor synthesis 1 (ENSG00000124615), score: 0.58 MOGmyelin oligodendrocyte glycoprotein (ENSG00000204655), score: 0.58 MPLmyeloproliferative leukemia virus oncogene (ENSG00000117400), score: 0.56 MRPL39mitochondrial ribosomal protein L39 (ENSG00000154719), score: -0.55 MSGN1mesogenin 1 (ENSG00000151379), score: 0.62 MTERFD1MTERF domain containing 1 (ENSG00000156469), score: -0.56 MTMR10myotubularin related protein 10 (ENSG00000166912), score: 0.53 MYO9Bmyosin IXB (ENSG00000099331), score: 0.68 NAA38N(alpha)-acetyltransferase 38, NatC auxiliary subunit (ENSG00000128534), score: -0.53 NDST1N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (ENSG00000070614), score: 0.52 NEO1neogenin 1 (ENSG00000067141), score: 0.54 NIPA1non imprinted in Prader-Willi/Angelman syndrome 1 (ENSG00000170113), score: 0.57 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 0.88 NKX2-2NK2 homeobox 2 (ENSG00000125820), score: 0.64 NOL11nucleolar protein 11 (ENSG00000130935), score: -0.59 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: -0.61 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.92 NTNG2netrin G2 (ENSG00000196358), score: 0.55 NUCB2nucleobindin 2 (ENSG00000070081), score: -0.54 OLIG2oligodendrocyte lineage transcription factor 2 (ENSG00000205927), score: 0.59 OPALINoligodendrocytic myelin paranodal and inner loop protein (ENSG00000197430), score: 0.57 OXSM3-oxoacyl-ACP synthase, mitochondrial (ENSG00000151093), score: -0.57 P2RX7purinergic receptor P2X, ligand-gated ion channel, 7 (ENSG00000089041), score: 0.59 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: 0.61 PAQR6progestin and adipoQ receptor family member VI (ENSG00000160781), score: 0.58 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.57 PGM2phosphoglucomutase 2 (ENSG00000169299), score: -0.59 PHLDA3pleckstrin homology-like domain, family A, member 3 (ENSG00000174307), score: 0.55 PHLDB1pleckstrin homology-like domain, family B, member 1 (ENSG00000019144), score: 0.55 PHLPP1PH domain and leucine rich repeat protein phosphatase 1 (ENSG00000081913), score: 0.63 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (ENSG00000171608), score: 0.64 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.7 PLBD2phospholipase B domain containing 2 (ENSG00000151176), score: 0.54 PLEKHB1pleckstrin homology domain containing, family B (evectins) member 1 (ENSG00000021300), score: 0.55 PLEKHG3pleckstrin homology domain containing, family G (with RhoGef domain) member 3 (ENSG00000126822), score: 0.63 PLEKHH1pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 (ENSG00000054690), score: 0.52 PLLPplasmolipin (ENSG00000102934), score: 0.56 PLP1proteolipid protein 1 (ENSG00000123560), score: 0.52 PLXNB3plexin B3 (ENSG00000198753), score: 0.61 PMP22peripheral myelin protein 22 (ENSG00000109099), score: 0.55 PNPLA6patatin-like phospholipase domain containing 6 (ENSG00000032444), score: 0.55 POLR2Gpolymerase (RNA) II (DNA directed) polypeptide G (ENSG00000168002), score: 0.55 POMGNT1protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase (ENSG00000085998), score: 0.55 PPP1R14Aprotein phosphatase 1, regulatory (inhibitor) subunit 14A (ENSG00000167641), score: 0.6 PRAF2PRA1 domain family, member 2 (ENSG00000243279), score: 0.53 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.72 PRKCQprotein kinase C, theta (ENSG00000065675), score: 0.67 PRMT2protein arginine methyltransferase 2 (ENSG00000160310), score: 0.53 PROKR2prokineticin receptor 2 (ENSG00000101292), score: 0.52 PRPF18PRP18 pre-mRNA processing factor 18 homolog (S. cerevisiae) (ENSG00000165630), score: -0.68 PRPH2peripherin 2 (retinal degeneration, slow) (ENSG00000112619), score: 0.57 PRR18proline rich 18 (ENSG00000176381), score: 0.8 PRR5Lproline rich 5 like (ENSG00000135362), score: 0.65 PRR7proline rich 7 (synaptic) (ENSG00000131188), score: 0.53 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: 0.76 PSRC1proline/serine-rich coiled-coil 1 (ENSG00000134222), score: 0.56 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: 0.68 PTPRHprotein tyrosine phosphatase, receptor type, H (ENSG00000080031), score: 0.82 PXKPX domain containing serine/threonine kinase (ENSG00000168297), score: 0.63 RAB11BRAB11B, member RAS oncogene family (ENSG00000185236), score: 0.59 RAB4BRAB4B, member RAS oncogene family (ENSG00000167578), score: 0.7 RALGDSral guanine nucleotide dissociation stimulator (ENSG00000160271), score: 0.58 RAPGEF5Rap guanine nucleotide exchange factor (GEF) 5 (ENSG00000136237), score: 0.6 RARSarginyl-tRNA synthetase (ENSG00000113643), score: -0.57 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.62 RBP3retinol binding protein 3, interstitial (ENSG00000107618), score: 0.55 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (ENSG00000158315), score: 0.58 RHOrhodopsin (ENSG00000163914), score: 0.6 RHOGras homolog gene family, member G (rho G) (ENSG00000177105), score: 0.72 RHOUras homolog gene family, member U (ENSG00000116574), score: 0.61 RIOK2RIO kinase 2 (yeast) (ENSG00000058729), score: -0.63 RNF220ring finger protein 220 (ENSG00000187147), score: 0.53 ROBO3roundabout, axon guidance receptor, homolog 3 (Drosophila) (ENSG00000154134), score: 0.55 ROM1retinal outer segment membrane protein 1 (ENSG00000149489), score: 0.69 RPF1ribosome production factor 1 homolog (S. cerevisiae) (ENSG00000117133), score: -0.64 RPP30ribonuclease P/MRP 30kDa subunit (ENSG00000148688), score: -0.54 RS1retinoschisin 1 (ENSG00000102104), score: 0.66 SAMD4Bsterile alpha motif domain containing 4B (ENSG00000179134), score: 0.61 SCNN1Dsodium channel, nonvoltage-gated 1, delta (ENSG00000162572), score: 0.6 SCRIBscribbled homolog (Drosophila) (ENSG00000180900), score: 0.57 SEC14L5SEC14-like 5 (S. cerevisiae) (ENSG00000103184), score: 0.56 SEMA3Bsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B (ENSG00000012171), score: 0.57 SEMA4Csema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C (ENSG00000168758), score: 0.54 SEMA4Dsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D (ENSG00000187764), score: 0.69 SEMA6Asema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A (ENSG00000092421), score: 0.64 SETD6SET domain containing 6 (ENSG00000103037), score: -0.61 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.57 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: 0.69 SH3TC2SH3 domain and tetratricopeptide repeats 2 (ENSG00000169247), score: 0.73 SIRT2sirtuin 2 (ENSG00000068903), score: 0.75 SLAIN1SLAIN motif family, member 1 (ENSG00000139737), score: 0.59 SLC22A15solute carrier family 22, member 15 (ENSG00000163393), score: 0.56 SLC44A1solute carrier family 44, member 1 (ENSG00000070214), score: 0.6 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.65 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.56 SLCO1A2solute carrier organic anion transporter family, member 1A2 (ENSG00000084453), score: 0.61 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.6 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (ENSG00000100796), score: -0.54 SNAI3snail homolog 3 (Drosophila) (ENSG00000185669), score: 0.53 SNX2sorting nexin 2 (ENSG00000205302), score: -0.53 SOAT1sterol O-acyltransferase 1 (ENSG00000057252), score: -0.53 SORCS2sortilin-related VPS10 domain containing receptor 2 (ENSG00000184985), score: 0.56 SOX10SRY (sex determining region Y)-box 10 (ENSG00000100146), score: 0.61 SOX8SRY (sex determining region Y)-box 8 (ENSG00000005513), score: 0.63 SRPRBsignal recognition particle receptor, B subunit (ENSG00000144867), score: 0.57 SSBSjogren syndrome antigen B (autoantigen La) (ENSG00000138385), score: -0.61 ST7suppression of tumorigenicity 7 (ENSG00000004866), score: -0.55 SUZ12suppressor of zeste 12 homolog (Drosophila) (ENSG00000178691), score: -0.53 SYNJ2synaptojanin 2 (ENSG00000078269), score: 0.56 SYT15synaptotagmin XV (ENSG00000204176), score: 0.67 TBC1D2TBC1 domain family, member 2 (ENSG00000095383), score: 0.53 TBC1D9BTBC1 domain family, member 9B (with GRAM domain) (ENSG00000197226), score: 0.53 TBCBtubulin folding cofactor B (ENSG00000105254), score: 0.65 TBCEtubulin folding cofactor E (ENSG00000116957), score: -0.67 TGFAtransforming growth factor, alpha (ENSG00000163235), score: 0.53 THBS2thrombospondin 2 (ENSG00000186340), score: 0.58 THOC2THO complex 2 (ENSG00000125676), score: -0.53 THRAP3thyroid hormone receptor associated protein 3 (ENSG00000054118), score: -0.53 THUMPD1THUMP domain containing 1 (ENSG00000066654), score: -0.59 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: 0.58 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.74 TMCC3transmembrane and coiled-coil domain family 3 (ENSG00000057704), score: 0.58 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.76 TMEM151Atransmembrane protein 151A (ENSG00000179292), score: 0.61 TMEM175transmembrane protein 175 (ENSG00000127419), score: 0.54 TMEM184Btransmembrane protein 184B (ENSG00000198792), score: 0.52 TMEM199transmembrane protein 199 (ENSG00000244045), score: -0.55 TMIEtransmembrane inner ear (ENSG00000181585), score: 0.52 TMPRSS5transmembrane protease, serine 5 (ENSG00000166682), score: 0.61 TNFRSF13Ctumor necrosis factor receptor superfamily, member 13C (ENSG00000159958), score: 0.65 TP53INP2tumor protein p53 inducible nuclear protein 2 (ENSG00000078804), score: 0.7 TRAPPC10trafficking protein particle complex 10 (ENSG00000160218), score: 0.65 TREM2triggering receptor expressed on myeloid cells 2 (ENSG00000095970), score: 0.54 TRIM41tripartite motif-containing 41 (ENSG00000146063), score: 0.65 TSPAN15tetraspanin 15 (ENSG00000099282), score: 0.56 UGT8UDP glycosyltransferase 8 (ENSG00000174607), score: 0.6 UNC5Cunc-5 homolog C (C. elegans) (ENSG00000182168), score: 0.74 USP54ubiquitin specific peptidase 54 (ENSG00000166348), score: 0.54 UTS2urotensin 2 (ENSG00000049247), score: 0.52 VSTM2BV-set and transmembrane domain containing 2B (ENSG00000187135), score: 0.52 YME1L1YME1-like 1 (S. cerevisiae) (ENSG00000136758), score: -0.61 ZCCHC24zinc finger, CCHC domain containing 24 (ENSG00000165424), score: 0.59 ZEB2zinc finger E-box binding homeobox 2 (ENSG00000169554), score: 0.56 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.55 ZNF71zinc finger protein 71 (ENSG00000197951), score: 0.54 ZSWIM6zinc finger, SWIM-type containing 6 (ENSG00000130449), score: 0.66
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |