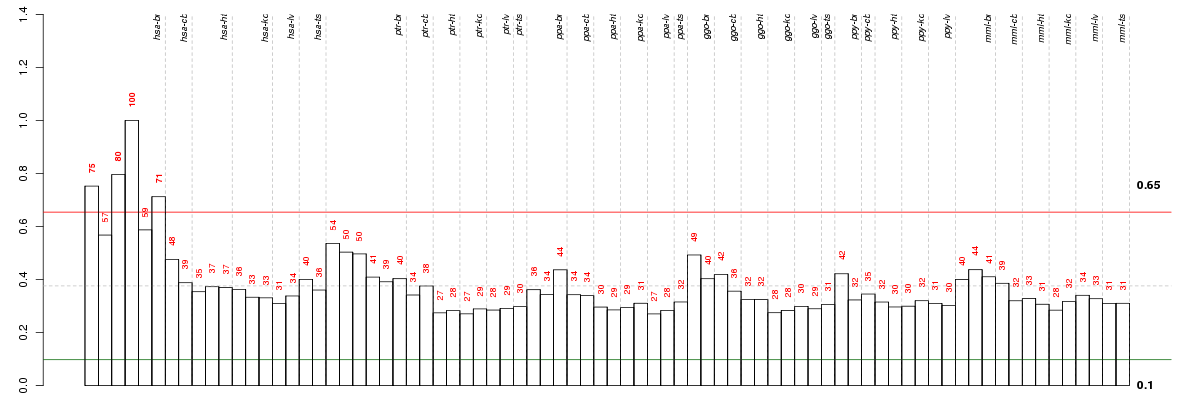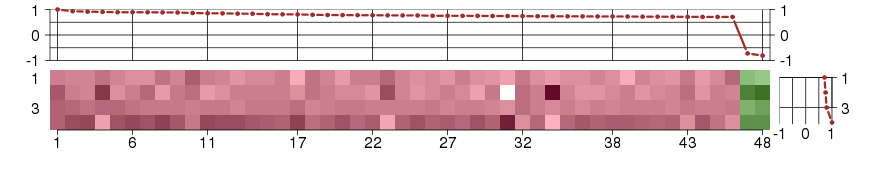

Under-expression is coded with green,
over-expression with red color.



ATP10BATPase, class V, type 10B (ENSG00000118322), score: 0.72 C1orf198chromosome 1 open reading frame 198 (ENSG00000119280), score: 0.73 C3orf16chromosome 3 open reading frame 16 (ENSG00000206199), score: 0.71 CCT8chaperonin containing TCP1, subunit 8 (theta) (ENSG00000156261), score: -0.81 CD22CD22 molecule (ENSG00000012124), score: 0.91 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 0.88 CHADLchondroadherin-like (ENSG00000100399), score: 0.82 CLCA4chloride channel accessory 4 (ENSG00000016602), score: 0.85 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.73 CLN8ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) (ENSG00000182372), score: 0.89 CLUL1clusterin-like 1 (retinal) (ENSG00000079101), score: 0.86 CNP2',3'-cyclic nucleotide 3' phosphodiesterase (ENSG00000173786), score: 0.77 CRYBB1crystallin, beta B1 (ENSG00000100122), score: 0.71 CYB5D1cytochrome b5 domain containing 1 (ENSG00000182224), score: -0.72 DEFB134defensin, beta 134 (ENSG00000205882), score: 0.77 DMBT1deleted in malignant brain tumors 1 (ENSG00000187908), score: 0.81 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.78 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.78 FABP6fatty acid binding protein 6, ileal (ENSG00000170231), score: 0.74 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.83 FAM125Bfamily with sequence similarity 125, member B (ENSG00000196814), score: 0.72 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (ENSG00000139629), score: 0.89 GIPRgastric inhibitory polypeptide receptor (ENSG00000010310), score: 0.74 GPR78G protein-coupled receptor 78 (ENSG00000155269), score: 0.73 GREM1gremlin 1 (ENSG00000166923), score: 0.89 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.75 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.73 KREMEN2kringle containing transmembrane protein 2 (ENSG00000131650), score: 1 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.85 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (ENSG00000107902), score: 0.75 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.79 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.91 MARCKSL1MARCKS-like 1 (ENSG00000175130), score: 0.74 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 0.89 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.93 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.72 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.73 PRR18proline rich 18 (ENSG00000176381), score: 0.81 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: 0.78 PTPRHprotein tyrosine phosphatase, receptor type, H (ENSG00000080031), score: 0.84 RAB4BRAB4B, member RAS oncogene family (ENSG00000167578), score: 0.71 RHOGras homolog gene family, member G (rho G) (ENSG00000177105), score: 0.72 SH3TC2SH3 domain and tetratricopeptide repeats 2 (ENSG00000169247), score: 0.75 SIRT2sirtuin 2 (ENSG00000068903), score: 0.77 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.75 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.78 TP53INP2tumor protein p53 inducible nuclear protein 2 (ENSG00000078804), score: 0.71 UNC5Cunc-5 homolog C (C. elegans) (ENSG00000182168), score: 0.75
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |