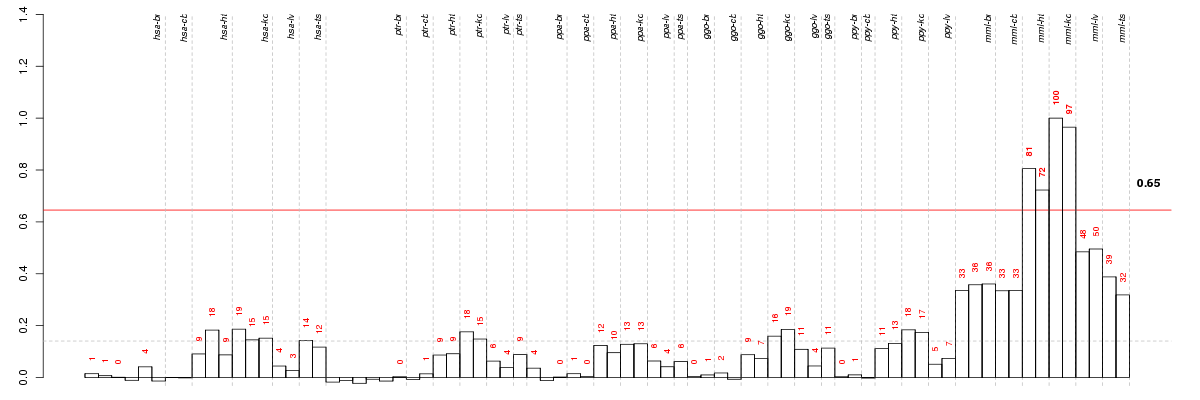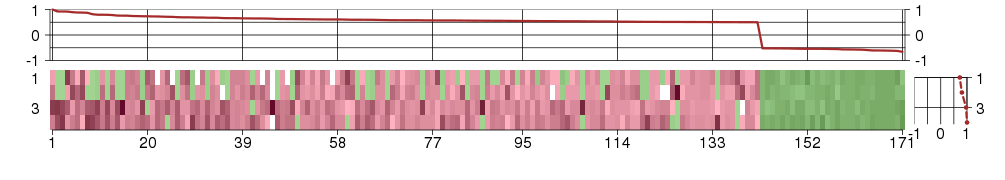

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively and non-covalently with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
hormone activity
The action characteristic of a hormone, any substance formed in very small amounts in one specialized organ or group of cells and carried (sometimes in the bloodstream) to another organ or group of cells in the same organism, upon which it has a specific regulatory action. The term was originally applied to agents with a stimulatory physiological action in vertebrate animals (as opposed to a chalone, which has a depressant action). Usage is now extended to regulatory compounds in lower animals and plants, and to synthetic substances having comparable effects; all bind receptors and trigger some biological process.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
amine transmembrane transporter activity
Catalysis of the transfer of amines, including polyamines, from one side of the membrane to the other. Amines are organic compounds that are weakly basic in character and contain an amino (-NH2) or substituted amino group.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
monoamine transmembrane transporter activity
Catalysis of the transfer of monoamines, organic compounds that contain one amino group that is connected to an aromatic ring by an ethylene group (-CH2-CH2-), from one side of the membrane to the other.
active transmembrane transporter activity
Catalysis of the transfer of a specific substance or related group of substances from one side of a membrane to the other, up the solute's concentration gradient. The transporter binds the solute and undergoes a series of conformational changes. Transport works equally well in either direction.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
amine transmembrane transporter activity
Catalysis of the transfer of amines, including polyamines, from one side of the membrane to the other. Amines are organic compounds that are weakly basic in character and contain an amino (-NH2) or substituted amino group.
ABI3BPABI family, member 3 (NESH) binding protein (ENSG00000154175), score: 0.5 ADIPOQadiponectin, C1Q and collagen domain containing (ENSG00000181092), score: 0.89 AGR3anterior gradient homolog 3 (Xenopus laevis) (ENSG00000173467), score: 0.79 AKR1CL1aldo-keto reductase family 1, member C-like 1 (ENSG00000196326), score: 0.65 ALOX12arachidonate 12-lipoxygenase (ENSG00000108839), score: 0.61 AMELXamelogenin, X-linked (ENSG00000125363), score: 0.64 ANGPTL7angiopoietin-like 7 (ENSG00000171819), score: 0.74 AP3S1adaptor-related protein complex 3, sigma 1 subunit (ENSG00000177879), score: -0.54 ATP10DATPase, class V, type 10D (ENSG00000145246), score: 0.51 ATP12AATPase, H+/K+ transporting, nongastric, alpha polypeptide (ENSG00000075673), score: 0.6 B3GAT2beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) (ENSG00000112309), score: -0.63 BCCIPBRCA2 and CDKN1A interacting protein (ENSG00000107949), score: 0.5 BIRC7baculoviral IAP repeat-containing 7 (ENSG00000101197), score: 0.65 BMP3bone morphogenetic protein 3 (ENSG00000152785), score: 0.71 BTCbetacellulin (ENSG00000174808), score: 0.68 C14orf138chromosome 14 open reading frame 138 (ENSG00000100483), score: -0.54 C15orf57chromosome 15 open reading frame 57 (ENSG00000128891), score: -0.54 C16orf91chromosome 16 open reading frame 91 (ENSG00000174109), score: 0.59 C17orf49chromosome 17 open reading frame 49 (ENSG00000161939), score: 0.52 C17orf78chromosome 17 open reading frame 78 (ENSG00000167230), score: 0.74 C22orf15chromosome 22 open reading frame 15 (ENSG00000169314), score: 0.66 C22orf39chromosome 22 open reading frame 39 (ENSG00000242259), score: -0.66 C6orf52chromosome 6 open reading frame 52 (ENSG00000137434), score: 0.62 CA13carbonic anhydrase XIII (ENSG00000185015), score: 0.57 CABP5calcium binding protein 5 (ENSG00000105507), score: 0.61 CCDC142coiled-coil domain containing 142 (ENSG00000135637), score: 0.53 CCL13chemokine (C-C motif) ligand 13 (ENSG00000181374), score: 0.73 CCL25chemokine (C-C motif) ligand 25 (ENSG00000131142), score: 0.55 CCR3chemokine (C-C motif) receptor 3 (ENSG00000183625), score: 0.71 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.57 CD164L2CD164 sialomucin-like 2 (ENSG00000174950), score: 0.63 CD3GCD3g molecule, gamma (CD3-TCR complex) (ENSG00000160654), score: 0.63 CDK5cyclin-dependent kinase 5 (ENSG00000164885), score: -0.53 CDV3CDV3 homolog (mouse) (ENSG00000091527), score: -0.59 CHIC2cysteine-rich hydrophobic domain 2 (ENSG00000109220), score: 0.55 CHRNA1cholinergic receptor, nicotinic, alpha 1 (muscle) (ENSG00000138435), score: 0.5 CLINT1clathrin interactor 1 (ENSG00000113282), score: 0.51 COCHcoagulation factor C homolog, cochlin (Limulus polyphemus) (ENSG00000100473), score: 0.72 COL4A6collagen, type IV, alpha 6 (ENSG00000197565), score: 0.65 CPA3carboxypeptidase A3 (mast cell) (ENSG00000163751), score: 0.57 CSN2casein beta (ENSG00000135222), score: 0.76 DCBLD1discoidin, CUB and LCCL domain containing 1 (ENSG00000164465), score: -0.55 DEFB4Bdefensin, beta 4B (ENSG00000171711), score: 0.92 DISP1dispatched homolog 1 (Drosophila) (ENSG00000154309), score: 0.57 ELMOD3ELMO/CED-12 domain containing 3 (ENSG00000115459), score: 0.58 ESM1endothelial cell-specific molecule 1 (ENSG00000164283), score: 0.61 FAM120AOSfamily with sequence similarity 120A opposite strand (ENSG00000188938), score: 0.61 FBXW8F-box and WD repeat domain containing 8 (ENSG00000174989), score: 0.51 FGFBP1fibroblast growth factor binding protein 1 (ENSG00000137440), score: 0.73 FKTNfukutin (ENSG00000106692), score: 0.51 FNDC1fibronectin type III domain containing 1 (ENSG00000164694), score: 0.55 FOXN1forkhead box N1 (ENSG00000109101), score: 0.54 FRZBfrizzled-related protein (ENSG00000162998), score: 0.56 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.6 GALgalanin prepropeptide (ENSG00000069482), score: 0.62 GFRALGDNF family receptor alpha like (ENSG00000187871), score: 0.68 GPR1G protein-coupled receptor 1 (ENSG00000183671), score: 0.69 GPR113G protein-coupled receptor 113 (ENSG00000173567), score: 0.53 GPRC6AG protein-coupled receptor, family C, group 6, member A (ENSG00000173612), score: 0.6 GPX8glutathione peroxidase 8 (putative) (ENSG00000164294), score: 0.58 GTF2F1general transcription factor IIF, polypeptide 1, 74kDa (ENSG00000125651), score: -0.57 HNRNPLheterogeneous nuclear ribonucleoprotein L (ENSG00000104824), score: -0.62 HOXA2homeobox A2 (ENSG00000105996), score: 0.57 HOXB1homeobox B1 (ENSG00000120094), score: 0.53 HOXB6homeobox B6 (ENSG00000108511), score: 0.5 HTR1D5-hydroxytryptamine (serotonin) receptor 1D (ENSG00000179546), score: 0.6 IFIT1Binterferon-induced protein with tetratricopeptide repeats 1B (ENSG00000204010), score: 0.75 IGFBP6insulin-like growth factor binding protein 6 (ENSG00000167779), score: 0.52 IGFL3IGF-like family member 3 (ENSG00000188624), score: 0.52 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.69 INO80EINO80 complex subunit E (ENSG00000169592), score: -0.61 KCTD7potassium channel tetramerisation domain containing 7 (ENSG00000154710), score: -0.55 KIAA1609KIAA1609 (ENSG00000140950), score: 0.56 KIAA1841KIAA1841 (ENSG00000162929), score: -0.54 KLF8Kruppel-like factor 8 (ENSG00000102349), score: 0.57 KLHL13kelch-like 13 (Drosophila) (ENSG00000003096), score: 0.53 KRT3keratin 3 (ENSG00000186442), score: 0.76 KRTAP3-1keratin associated protein 3-1 (ENSG00000212901), score: 0.65 LAMB4laminin, beta 4 (ENSG00000091128), score: 0.81 LEPleptin (ENSG00000174697), score: 0.88 LGALS12lectin, galactoside-binding, soluble, 12 (ENSG00000133317), score: 0.5 LOC100127905family with sequence similarity 165, member B pseudogene (ENSG00000198738), score: 0.76 LRRC14leucine rich repeat containing 14 (ENSG00000160959), score: -0.52 LUC7LLUC7-like (S. cerevisiae) (ENSG00000007392), score: -0.52 MAPK7mitogen-activated protein kinase 7 (ENSG00000166484), score: 0.6 MAT2Bmethionine adenosyltransferase II, beta (ENSG00000038274), score: 0.56 MATN3matrilin 3 (ENSG00000132031), score: 0.61 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.57 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.57 MRPL35mitochondrial ribosomal protein L35 (ENSG00000132313), score: 0.55 MS4A10membrane-spanning 4-domains, subfamily A, member 10 (ENSG00000172689), score: 0.63 MS4A8Bmembrane-spanning 4-domains, subfamily A, member 8B (ENSG00000166959), score: 0.92 MSLNmesothelin (ENSG00000102854), score: 0.58 MSLNLmesothelin-like (ENSG00000162006), score: 1 MTMR8myotubularin related protein 8 (ENSG00000102043), score: 0.54 MXRA5matrix-remodelling associated 5 (ENSG00000101825), score: 0.54 MYL1myosin, light chain 1, alkali; skeletal, fast (ENSG00000168530), score: 0.6 NAA40N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) (ENSG00000110583), score: -0.61 NET1neuroepithelial cell transforming 1 (ENSG00000173848), score: 0.54 NGRNneugrin, neurite outgrowth associated (ENSG00000182768), score: -0.61 NME6non-metastatic cells 6, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000172113), score: 0.51 NOP14NOP14 nucleolar protein homolog (yeast) (ENSG00000087269), score: 0.61 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (ENSG00000113389), score: 0.54 NUDT15nudix (nucleoside diphosphate linked moiety X)-type motif 15 (ENSG00000136159), score: -0.55 OLFML1olfactomedin-like 1 (ENSG00000183801), score: 0.52 OR10Q1olfactory receptor, family 10, subfamily Q, member 1 (ENSG00000180475), score: 0.7 OTUD1OTU domain containing 1 (ENSG00000165312), score: 0.5 PAPLNpapilin, proteoglycan-like sulfated glycoprotein (ENSG00000100767), score: 0.54 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (ENSG00000102981), score: -0.52 PARP15poly (ADP-ribose) polymerase family, member 15 (ENSG00000173200), score: 0.58 PHOX2Bpaired-like homeobox 2b (ENSG00000109132), score: 0.68 PLA2G2Dphospholipase A2, group IID (ENSG00000117215), score: 0.52 PLA2G3phospholipase A2, group III (ENSG00000100078), score: 0.8 PLEKHG7pleckstrin homology domain containing, family G (with RhoGef domain) member 7 (ENSG00000187510), score: 0.57 PPYpancreatic polypeptide (ENSG00000108849), score: 0.79 PTCD2pentatricopeptide repeat domain 2 (ENSG00000049883), score: 0.5 PTPLAD2protein tyrosine phosphatase-like A domain containing 2 (ENSG00000188921), score: 0.51 QRSL1glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 (ENSG00000130348), score: 0.51 RAB1BRAB1B, member RAS oncogene family (ENSG00000174903), score: -0.57 RABEPKRab9 effector protein with kelch motifs (ENSG00000136933), score: -0.58 ROR1receptor tyrosine kinase-like orphan receptor 1 (ENSG00000185483), score: 0.51 RSL1D1ribosomal L1 domain containing 1 (ENSG00000171490), score: 0.55 RTN4IP1reticulon 4 interacting protein 1 (ENSG00000130347), score: 0.53 SAC3D1SAC3 domain containing 1 (ENSG00000168061), score: -0.54 SBK2SH3-binding domain kinase family, member 2 (ENSG00000187550), score: 0.51 SCLT1sodium channel and clathrin linker 1 (ENSG00000151466), score: 0.65 SEPT4septin 4 (ENSG00000108387), score: -0.53 SERINC4serine incorporator 4 (ENSG00000184716), score: 0.68 SETDB2SET domain, bifurcated 2 (ENSG00000136169), score: 0.69 SFRP4secreted frizzled-related protein 4 (ENSG00000106483), score: 0.56 SH2D7SH2 domain containing 7 (ENSG00000183476), score: 0.51 SHISA2shisa homolog 2 (Xenopus laevis) (ENSG00000180730), score: 0.66 SIP1survival of motor neuron protein interacting protein 1 (ENSG00000092208), score: -0.57 SLC15A5solute carrier family 15, member 5 (ENSG00000188991), score: 0.87 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.52 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.58 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.54 SLC5A7solute carrier family 5 (choline transporter), member 7 (ENSG00000115665), score: 0.62 SLC6A3solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 (ENSG00000142319), score: 0.56 SLC6A4solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 (ENSG00000108576), score: 0.88 SLC7A10solute carrier family 7, (neutral amino acid transporter, y+ system) member 10 (ENSG00000130876), score: 0.55 SOX14SRY (sex determining region Y)-box 14 (ENSG00000168875), score: 0.64 SRP14signal recognition particle 14kDa (homologous Alu RNA binding protein) (ENSG00000140319), score: -0.61 STRA13stimulated by retinoic acid 13 homolog (mouse) (ENSG00000169689), score: -0.53 TAAR2trace amine associated receptor 2 (ENSG00000146378), score: 0.56 TBX19T-box 19 (ENSG00000143178), score: 0.63 TCEA2transcription elongation factor A (SII), 2 (ENSG00000171703), score: -0.54 TFECtranscription factor EC (ENSG00000105967), score: 0.55 TMEM167Btransmembrane protein 167B (ENSG00000215717), score: 0.51 TMEM43transmembrane protein 43 (ENSG00000170876), score: 0.61 TMIGD1transmembrane and immunoglobulin domain containing 1 (ENSG00000182271), score: 0.66 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.66 TNFSF15tumor necrosis factor (ligand) superfamily, member 15 (ENSG00000181634), score: 0.58 TRMT1TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae) (ENSG00000104907), score: -0.53 UCMAupper zone of growth plate and cartilage matrix associated (ENSG00000165623), score: 0.51 UCN3urocortin 3 (stresscopin) (ENSG00000178473), score: 0.92 UPK1Buroplakin 1B (ENSG00000114638), score: 0.56 UPK2uroplakin 2 (ENSG00000110375), score: 0.68 XGXg blood group (ENSG00000124343), score: 0.5 XPCxeroderma pigmentosum, complementation group C (ENSG00000154767), score: 0.57 ZNF114zinc finger protein 114 (ENSG00000178150), score: 0.59 ZNF146zinc finger protein 146 (ENSG00000167635), score: -0.57 ZNF16zinc finger protein 16 (ENSG00000170631), score: 0.56 ZNF805zinc finger protein 805 (ENSG00000204524), score: 0.52 ZP4zona pellucida glycoprotein 4 (ENSG00000116996), score: 0.51 ZSWIM3zinc finger, SWIM-type containing 3 (ENSG00000132801), score: 0.52
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_ht_f_ca1 | mml | ht | f | _ |
| mml_ht_m_ca1 | mml | ht | m | _ |
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |