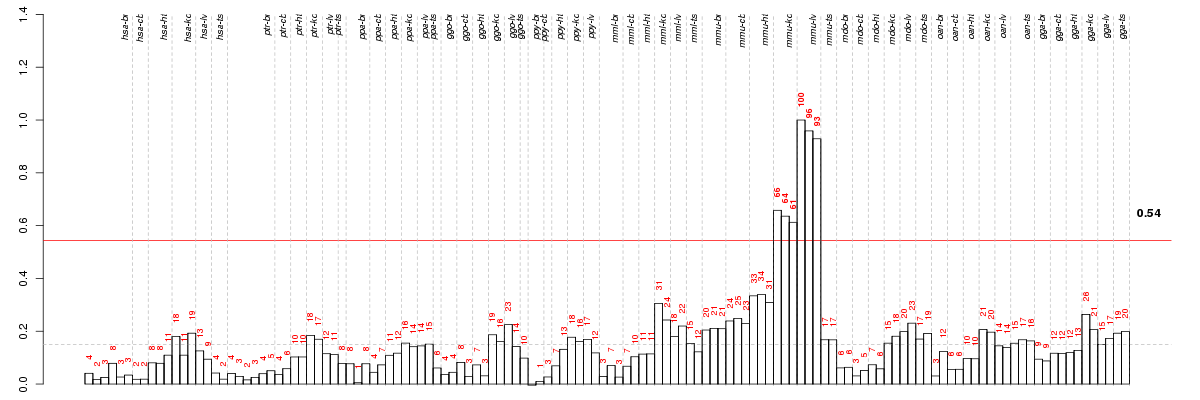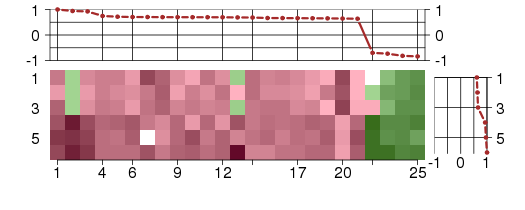

Under-expression is coded with green,
over-expression with red color.



ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.72 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.82 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.7 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: 0.7 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.75 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.69 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.66 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.92 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.7 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.67 FABP2fatty acid binding protein 2, intestinal (ENSG00000145384), score: 0.94 GJB2gap junction protein, beta 2, 26kDa (ENSG00000165474), score: 0.66 H1FXH1 histone family, member X (ENSG00000184897), score: -0.84 LIFRleukemia inhibitory factor receptor alpha (ENSG00000113594), score: 0.7 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.64 PLK3polo-like kinase 3 (ENSG00000173846), score: 0.65 PRKD3protein kinase D3 (ENSG00000115825), score: 0.66 PRLRprolactin receptor (ENSG00000113494), score: 0.7 SCCPDHsaccharopine dehydrogenase (putative) (ENSG00000143653), score: -0.73 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.68 SLC17A8solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 (ENSG00000179520), score: 0.69 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.65 TSPAN3tetraspanin 3 (ENSG00000140391), score: -0.7 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.7 ZNF750zinc finger protein 750 (ENSG00000141579), score: 1
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |