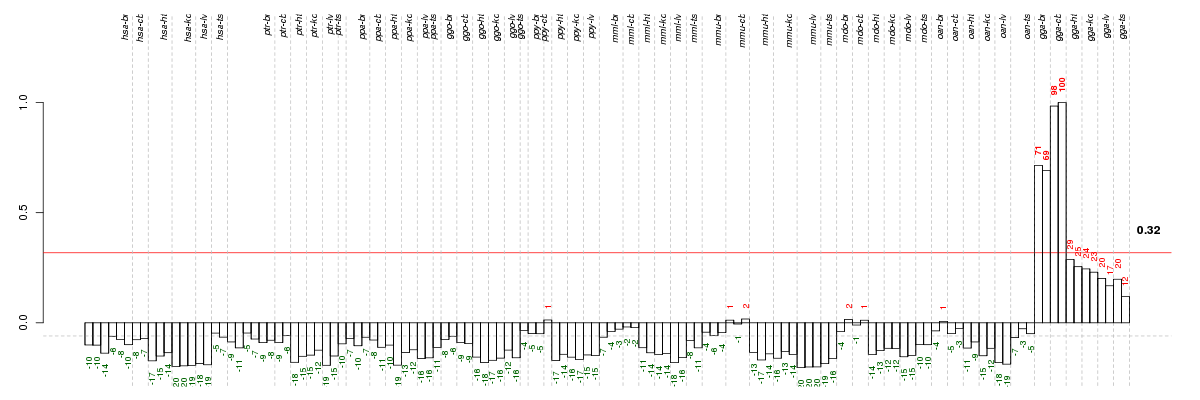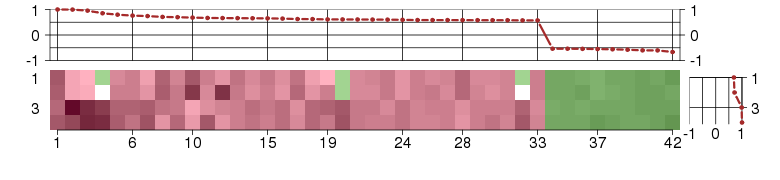

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 4.617e-02 | 0.5537 | 4 | 102 | Neuroactive ligand-receptor interaction |
ALX4ALX homeobox 4 (ENSG00000052850), score: 0.71 ANGPT2angiopoietin 2 (ENSG00000091879), score: 0.65 ATP13A5ATPase type 13A5 (ENSG00000187527), score: 0.63 BET3LBET3 like (S. cerevisiae) (ENSG00000173626), score: 0.85 BLKB lymphoid tyrosine kinase (ENSG00000136573), score: 0.74 BPIL3bactericidal/permeability-increasing protein-like 3 (ENSG00000167104), score: 0.6 C18orf22chromosome 18 open reading frame 22 (ENSG00000101546), score: -0.56 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.54 DOCK11dedicator of cytokinesis 11 (ENSG00000147251), score: 0.64 EN1engrailed homeobox 1 (ENSG00000163064), score: 0.61 EPGNepithelial mitogen homolog (mouse) (ENSG00000182585), score: 0.58 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (ENSG00000111886), score: 0.8 GFRALGDNF family receptor alpha like (ENSG00000187871), score: 0.67 GLRA4glycine receptor, alpha 4 (ENSG00000188828), score: 0.66 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 0.7 GRIK1glutamate receptor, ionotropic, kainate 1 (ENSG00000171189), score: 0.66 INPPL1inositol polyphosphate phosphatase-like 1 (ENSG00000165458), score: -0.6 KCNA10potassium voltage-gated channel, shaker-related subfamily, member 10 (ENSG00000143105), score: 0.96 KERAkeratocan (ENSG00000139330), score: 1 MCF2MCF.2 cell line derived transforming sequence (ENSG00000101977), score: 0.59 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.57 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: 0.58 MYOGmyogenin (myogenic factor 4) (ENSG00000122180), score: 1 NUSAP1nucleolar and spindle associated protein 1 (ENSG00000137804), score: 0.61 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.66 P2RY1purinergic receptor P2Y, G-protein coupled, 1 (ENSG00000169860), score: 0.58 PANX3pannexin 3 (ENSG00000154143), score: 0.58 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.58 PDCLphosducin-like (ENSG00000136940), score: 0.58 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (ENSG00000116711), score: 0.62 POU1F1POU class 1 homeobox 1 (ENSG00000064835), score: 0.68 PPYR1pancreatic polypeptide receptor 1 (ENSG00000204174), score: 0.57 RHOrhodopsin (ENSG00000163914), score: 0.61 SDK2sidekick homolog 2 (chicken) (ENSG00000069188), score: 0.58 SLBPstem-loop binding protein (ENSG00000163950), score: -0.67 SNX3sorting nexin 3 (ENSG00000112335), score: -0.55 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.6 SYPL1synaptophysin-like 1 (ENSG00000008282), score: -0.6 TT, brachyury homolog (mouse) (ENSG00000164458), score: 0.76 TLL1tolloid-like 1 (ENSG00000038295), score: 0.59 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.54
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_br_f_ca1 | gga | br | f | _ |
| gga_br_m_ca1 | gga | br | m | _ |
| gga_cb_m_ca1 | gga | cb | m | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |