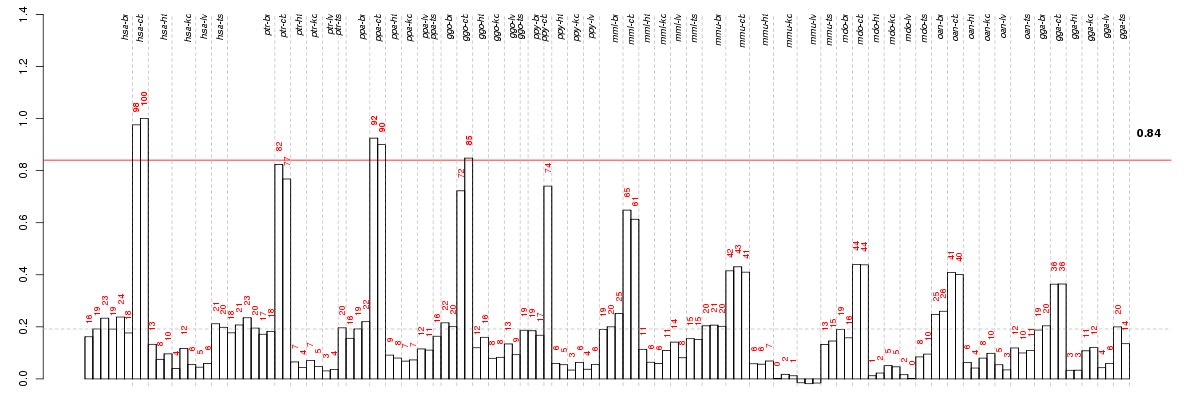

Under-expression is coded with green,
over-expression with red color.

response to tumor cell
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a tumor cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
NA


CCNJLcyclin J-like (ENSG00000135083), score: 0.67 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.72 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.85 ESPNLespin-like (ENSG00000144488), score: 0.89 ETV6ets variant 6 (ENSG00000139083), score: -0.69 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.71 FGF3fibroblast growth factor 3 (ENSG00000186895), score: 1 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.68 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.7 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.79 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.64 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 0.87 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.65 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.9 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.81 TFAP2Etranscription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) (ENSG00000116819), score: 0.72 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.66 TP73tumor protein p73 (ENSG00000078900), score: 0.83 VSX1visual system homeobox 1 (ENSG00000100987), score: 0.93 ZIC4Zic family member 4 (ENSG00000174963), score: 0.71 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.64
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| hsa_cb_m_ca1 | hsa | cb | m | _ |
| hsa_cb_f_ca1 | hsa | cb | f | _ |