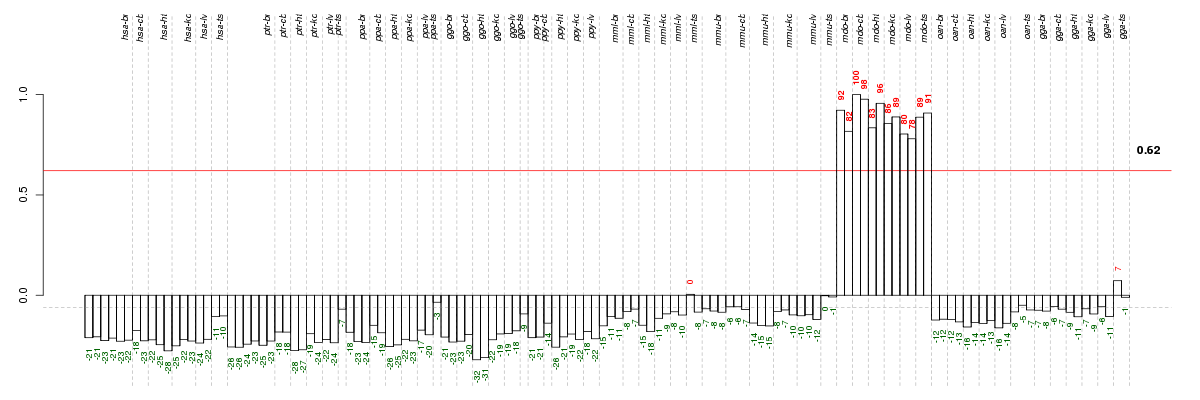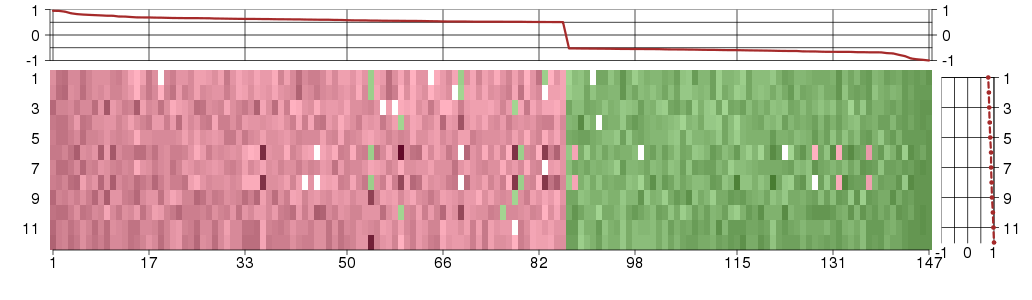

Under-expression is coded with green,
over-expression with red color.



AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: -0.61 AATFapoptosis antagonizing transcription factor (ENSG00000108270), score: -0.57 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: 0.59 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.51 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.68 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.63 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.62 AIREautoimmune regulator (ENSG00000160224), score: 0.52 ALKBH1alkB, alkylation repair homolog 1 (E. coli) (ENSG00000100601), score: 0.52 ALKBH2alkB, alkylation repair homolog 2 (E. coli) (ENSG00000189046), score: 0.63 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.6 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.51 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.64 ASB1ankyrin repeat and SOCS box-containing 1 (ENSG00000065802), score: 0.62 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (ENSG00000085978), score: 0.6 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.66 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.59 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.66 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.54 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: -0.57 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.68 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.69 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.65 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.6 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.76 C14orf101chromosome 14 open reading frame 101 (ENSG00000070269), score: -0.55 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.53 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: 0.61 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.63 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: -0.54 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.53 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.71 C3orf59chromosome 3 open reading frame 59 (ENSG00000180611), score: 0.52 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: 0.51 CA13carbonic anhydrase XIII (ENSG00000185015), score: 0.53 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.65 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: -0.64 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.79 CDK10cyclin-dependent kinase 10 (ENSG00000185324), score: -0.55 CLTBclathrin, light chain B (ENSG00000175416), score: -0.55 CNTFciliary neurotrophic factor (ENSG00000186660), score: 0.55 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.92 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.59 CYTBcytochrome b (ENSG00000198727), score: -1 CYTH1cytohesin 1 (ENSG00000108669), score: -0.67 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.58 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: 0.69 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: -0.55 DDX51DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 (ENSG00000185163), score: -0.53 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.66 ECDecdysoneless homolog (Drosophila) (ENSG00000122882), score: 0.62 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.66 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.6 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.56 EXOSC9exosome component 9 (ENSG00000123737), score: 0.55 FAM20Bfamily with sequence similarity 20, member B (ENSG00000116199), score: 0.51 FAM73Afamily with sequence similarity 73, member A (ENSG00000180488), score: 0.62 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.61 FBXO15F-box protein 15 (ENSG00000141665), score: 0.54 FBXO22F-box protein 22 (ENSG00000167196), score: 0.65 FBXO3F-box protein 3 (ENSG00000110429), score: 0.61 FOXN1forkhead box N1 (ENSG00000109101), score: 0.56 FRA10AC1fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 (ENSG00000148690), score: -0.57 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.85 GARSglycyl-tRNA synthetase (ENSG00000106105), score: 0.66 GEMIN5gem (nuclear organelle) associated protein 5 (ENSG00000082516), score: 0.51 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.64 GTF3Ageneral transcription factor IIIA (ENSG00000122034), score: -0.58 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (ENSG00000153187), score: 0.56 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.71 IKBIPIKBKB interacting protein (ENSG00000166130), score: 0.62 INVSinversin (ENSG00000119509), score: -0.59 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.95 IRAK1BP1interleukin-1 receptor-associated kinase 1 binding protein 1 (ENSG00000146243), score: 0.51 KBTBD8kelch repeat and BTB (POZ) domain containing 8 (ENSG00000163376), score: 0.57 KCTD5potassium channel tetramerisation domain containing 5 (ENSG00000167977), score: -0.55 KIAA0146KIAA0146 (ENSG00000164808), score: -0.54 KIAA0174KIAA0174 (ENSG00000182149), score: -0.63 KIAA1432KIAA1432 (ENSG00000107036), score: 0.52 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: 0.56 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: 0.54 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: -0.53 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.57 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.66 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: -0.61 MID2midline 2 (ENSG00000080561), score: 0.52 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.64 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.59 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.69 NARFnuclear prelamin A recognition factor (ENSG00000141562), score: -0.67 ND2MTND2 (ENSG00000198763), score: -0.95 ND6NADH dehydrogenase, subunit 6 (complex I) (ENSG00000198695), score: -0.62 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.63 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.92 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.72 OTOL1otolin 1 homolog (zebrafish) (ENSG00000182447), score: 0.57 PCID2PCI domain containing 2 (ENSG00000126226), score: -0.52 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.64 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.52 PGRprogesterone receptor (ENSG00000082175), score: 0.68 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.62 PHF21APHD finger protein 21A (ENSG00000135365), score: 0.6 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.56 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.63 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.72 PIGSphosphatidylinositol glycan anchor biosynthesis, class S (ENSG00000087111), score: 0.66 PODNpodocan (ENSG00000174348), score: 0.56 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.57 PRKACBprotein kinase, cAMP-dependent, catalytic, beta (ENSG00000142875), score: -0.57 PTRH2peptidyl-tRNA hydrolase 2 (ENSG00000141378), score: -0.53 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.75 RBPJLrecombination signal binding protein for immunoglobulin kappa J region-like (ENSG00000124232), score: 0.53 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: -0.58 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.78 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.67 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.53 SAP30LSAP30-like (ENSG00000164576), score: 0.66 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.55 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.55 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.66 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.95 SNRKSNF related kinase (ENSG00000163788), score: 0.55 SNX13sorting nexin 13 (ENSG00000071189), score: 0.66 SPG20spastic paraplegia 20 (Troyer syndrome) (ENSG00000133104), score: -0.6 SRPX2sushi-repeat-containing protein, X-linked 2 (ENSG00000102359), score: 0.58 STAMBPSTAM binding protein (ENSG00000124356), score: 0.52 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (ENSG00000040341), score: -0.54 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.67 STX8syntaxin 8 (ENSG00000170310), score: -0.59 TERTtelomerase reverse transcriptase (ENSG00000164362), score: 0.51 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.78 TMCO6transmembrane and coiled-coil domains 6 (ENSG00000113119), score: 0.57 TMEM138transmembrane protein 138 (ENSG00000149483), score: 0.53 TMEM18transmembrane protein 18 (ENSG00000151353), score: -0.68 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.8 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.77 TSNtranslin (ENSG00000211460), score: -0.66 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.83 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.55 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: -0.61 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.63 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.68 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: 0.52 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.97 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.82 ZNF800zinc finger protein 800 (ENSG00000048405), score: 0.52 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.72
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |