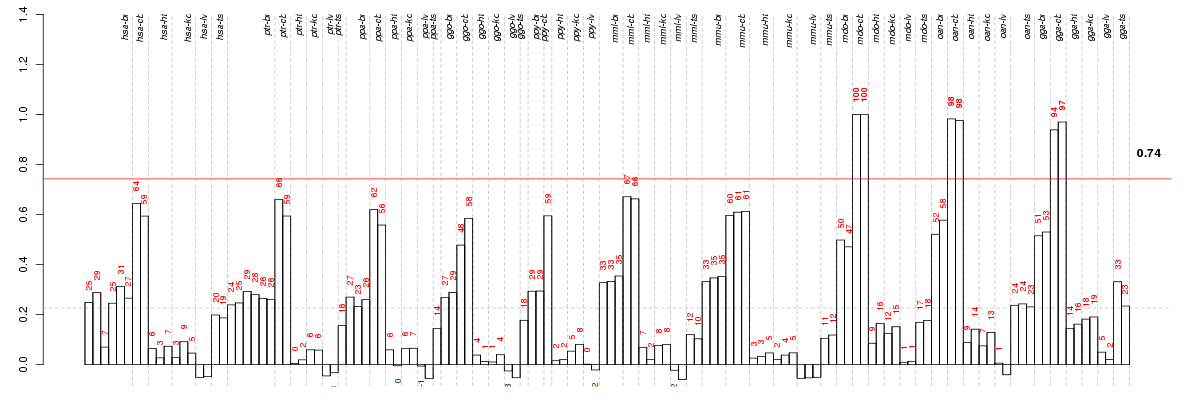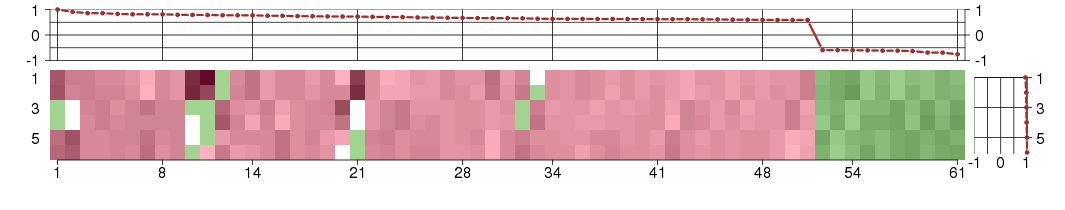

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
nerve-nerve synaptic transmission
The process of communication from a neuron to another neuron across a synapse.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
synaptic transmission, glutamatergic
The process of communication from a neuron to another neuron across a synapse using the neurotransmitter glutamate.
all
NA
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
ligand-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
ALG14asparagine-linked glycosylation 14 homolog (S. cerevisiae) (ENSG00000172339), score: -0.69 ASAMadipocyte-specific adhesion molecule (ENSG00000166250), score: 0.71 BET3LBET3 like (S. cerevisiae) (ENSG00000173626), score: 0.72 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.6 C14orf37chromosome 14 open reading frame 37 (ENSG00000139971), score: 0.59 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.63 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.63 CACHD1cache domain containing 1 (ENSG00000158966), score: 0.59 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.74 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.61 CHRNB4cholinergic receptor, nicotinic, beta 4 (ENSG00000117971), score: 0.77 CLVS2clavesin 2 (ENSG00000146352), score: 0.59 CNR1cannabinoid receptor 1 (brain) (ENSG00000118432), score: 0.63 CUL4Acullin 4A (ENSG00000139842), score: -0.6 EN1engrailed homeobox 1 (ENSG00000163064), score: 1 EPGNepithelial mitogen homolog (mouse) (ENSG00000182585), score: 0.91 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.79 FBXO22F-box protein 22 (ENSG00000167196), score: 0.62 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.75 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.71 FUBP1far upstream element (FUSE) binding protein 1 (ENSG00000162613), score: 0.63 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.61 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.62 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.76 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.86 GRIK1glutamate receptor, ionotropic, kainate 1 (ENSG00000171189), score: 0.66 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.67 KCNA10potassium voltage-gated channel, shaker-related subfamily, member 10 (ENSG00000143105), score: 0.79 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.68 KERAkeratocan (ENSG00000139330), score: 0.78 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: -0.6 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.73 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.63 LRRC38leucine rich repeat containing 38 (ENSG00000162494), score: 0.66 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.76 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.83 MAMLD1mastermind-like domain containing 1 (ENSG00000013619), score: 0.63 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.61 MIXL1Mix1 homeobox-like 1 (Xenopus laevis) (ENSG00000185155), score: 0.81 MTMR3myotubularin related protein 3 (ENSG00000100330), score: 0.6 NGBneuroglobin (ENSG00000165553), score: 0.7 OCIAD1OCIA domain containing 1 (ENSG00000109180), score: -0.59 PAX6paired box 6 (ENSG00000007372), score: 0.62 PTP4A2protein tyrosine phosphatase type IVA, member 2 (ENSG00000184007), score: -0.69 RXRAretinoid X receptor, alpha (ENSG00000186350), score: -0.62 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (ENSG00000197632), score: 0.72 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.65 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.69 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.64 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.63 SNRKSNF related kinase (ENSG00000163788), score: 0.66 SNX25sorting nexin 25 (ENSG00000109762), score: 0.73 SNX3sorting nexin 3 (ENSG00000112335), score: -0.6 TLL1tolloid-like 1 (ENSG00000038295), score: 0.77 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.77 TRPC3transient receptor potential cation channel, subfamily C, member 3 (ENSG00000138741), score: 0.86 UPF0639UPF0639 protein (ENSG00000175985), score: 0.81 ZIC4Zic family member 4 (ENSG00000174963), score: 0.81 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.59 ZNF385Czinc finger protein 385C (ENSG00000187595), score: 0.68 ZNF423zinc finger protein 423 (ENSG00000102935), score: 0.62
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_cb_m_ca1 | gga | cb | m | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |