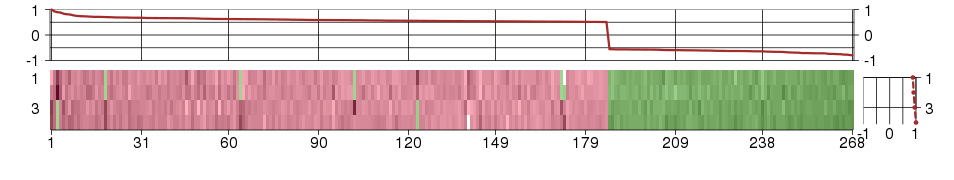

Under-expression is coded with green,
over-expression with red color.



AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.53 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.73 ACVR1Cactivin A receptor, type IC (ENSG00000123612), score: 0.57 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.75 ADPGKADP-dependent glucokinase (ENSG00000159322), score: 0.57 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.65 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.55 ALKBH1alkB, alkylation repair homolog 1 (E. coli) (ENSG00000100601), score: -0.61 AMDHD1amidohydrolase domain containing 1 (ENSG00000139344), score: 0.59 AMOTL1angiomotin like 1 (ENSG00000166025), score: -0.62 ANAPC2anaphase promoting complex subunit 2 (ENSG00000176248), score: -0.59 ANKLE2ankyrin repeat and LEM domain containing 2 (ENSG00000176915), score: -0.57 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.68 APLP2amyloid beta (A4) precursor-like protein 2 (ENSG00000084234), score: 0.54 AQRaquarius homolog (mouse) (ENSG00000021776), score: 0.57 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (ENSG00000101199), score: -0.58 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: 0.64 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (ENSG00000163947), score: -0.57 ASAH1N-acylsphingosine amidohydrolase (acid ceramidase) 1 (ENSG00000104763), score: 0.64 ASF1AASF1 anti-silencing function 1 homolog A (S. cerevisiae) (ENSG00000111875), score: 0.56 ASH2Lash2 (absent, small, or homeotic)-like (Drosophila) (ENSG00000129691), score: -0.6 ATF6activating transcription factor 6 (ENSG00000118217), score: 0.54 ATG13ATG13 autophagy related 13 homolog (S. cerevisiae) (ENSG00000175224), score: 0.54 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.68 AZI15-azacytidine induced 1 (ENSG00000141577), score: -0.62 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (ENSG00000158470), score: -0.63 BDKRB1bradykinin receptor B1 (ENSG00000100739), score: 0.81 BMP2bone morphogenetic protein 2 (ENSG00000125845), score: 0.54 C12orf26chromosome 12 open reading frame 26 (ENSG00000127720), score: 0.55 C14orf101chromosome 14 open reading frame 101 (ENSG00000070269), score: 0.59 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.64 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: -0.61 C18orf8chromosome 18 open reading frame 8 (ENSG00000141452), score: 0.62 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.51 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.61 C4orf34chromosome 4 open reading frame 34 (ENSG00000163683), score: 0.55 C5orf43chromosome 5 open reading frame 43 (ENSG00000188725), score: 0.53 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.69 C8orf40chromosome 8 open reading frame 40 (ENSG00000176209), score: -0.63 C9orf91chromosome 9 open reading frame 91 (ENSG00000157693), score: 0.56 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.62 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: 0.59 CCDC127coiled-coil domain containing 127 (ENSG00000164366), score: -0.69 CCNHcyclin H (ENSG00000134480), score: -0.67 CD82CD82 molecule (ENSG00000085117), score: 0.6 CD86CD86 molecule (ENSG00000114013), score: 0.68 CDC45cell division cycle 45 homolog (S. cerevisiae) (ENSG00000093009), score: 0.53 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.74 CHRNA9cholinergic receptor, nicotinic, alpha 9 (ENSG00000174343), score: 0.66 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.55 CIRBPcold inducible RNA binding protein (ENSG00000099622), score: -0.63 CLIP1CAP-GLY domain containing linker protein 1 (ENSG00000130779), score: -0.57 CLK4CDC-like kinase 4 (ENSG00000113240), score: -0.71 CLPXClpX caseinolytic peptidase X homolog (E. coli) (ENSG00000166855), score: 0.68 COG4component of oligomeric golgi complex 4 (ENSG00000103051), score: -0.57 COL4A3BPcollagen, type IV, alpha 3 (Goodpasture antigen) binding protein (ENSG00000113163), score: 0.65 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -0.77 COPB1coatomer protein complex, subunit beta 1 (ENSG00000129083), score: 0.55 CRLS1cardiolipin synthase 1 (ENSG00000088766), score: 0.55 CSE1LCSE1 chromosome segregation 1-like (yeast) (ENSG00000124207), score: -0.65 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.68 CXXC5CXXC finger 5 (ENSG00000171604), score: -0.64 CYP2R1cytochrome P450, family 2, subfamily R, polypeptide 1 (ENSG00000186104), score: 0.61 CYSLTR1cysteinyl leukotriene receptor 1 (ENSG00000173198), score: 0.63 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.62 DDA1DET1 and DDB1 associated 1 (ENSG00000130311), score: -0.58 DENND1BDENN/MADD domain containing 1B (ENSG00000213047), score: 0.62 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.63 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.58 DIRC2disrupted in renal carcinoma 2 (ENSG00000138463), score: 0.71 DNTTIP1deoxynucleotidyltransferase, terminal, interacting protein 1 (ENSG00000101457), score: 0.61 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.64 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.59 EDC3enhancer of mRNA decapping 3 homolog (S. cerevisiae) (ENSG00000179151), score: 0.56 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.69 EEF1E1eukaryotic translation elongation factor 1 epsilon 1 (ENSG00000124802), score: -0.69 EIF4E3eukaryotic translation initiation factor 4E family member 3 (ENSG00000163412), score: -0.61 EREGepiregulin (ENSG00000124882), score: 0.65 ERLEC1endoplasmic reticulum lectin 1 (ENSG00000068912), score: 0.6 ETNK2ethanolamine kinase 2 (ENSG00000143845), score: 0.51 EXOC8exocyst complex component 8 (ENSG00000116903), score: 0.63 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.57 FAM185Afamily with sequence similarity 185, member A (ENSG00000222011), score: -0.57 FAM3Cfamily with sequence similarity 3, member C (ENSG00000196937), score: -0.63 FAM63Bfamily with sequence similarity 63, member B (ENSG00000128923), score: 0.66 FAM83Bfamily with sequence similarity 83, member B (ENSG00000168143), score: 0.54 FBXO4F-box protein 4 (ENSG00000151876), score: 0.58 FGF19fibroblast growth factor 19 (ENSG00000162344), score: 0.52 FHfumarate hydratase (ENSG00000091483), score: 0.51 FSHBfollicle stimulating hormone, beta polypeptide (ENSG00000131808), score: 0.89 FTSJD2FtsJ methyltransferase domain containing 2 (ENSG00000137200), score: -0.6 FXR1fragile X mental retardation, autosomal homolog 1 (ENSG00000114416), score: -0.61 GALgalanin prepropeptide (ENSG00000069482), score: 0.62 GAR1GAR1 ribonucleoprotein homolog (yeast) (ENSG00000109534), score: 0.54 GAS6growth arrest-specific 6 (ENSG00000183087), score: -0.56 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.64 GGHgamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (ENSG00000137563), score: 0.52 GNMTglycine N-methyltransferase (ENSG00000124713), score: 0.52 GOLGA1golgin A1 (ENSG00000136935), score: -0.66 GOLIM4golgi integral membrane protein 4 (ENSG00000173905), score: 0.53 GPHNgephyrin (ENSG00000171723), score: 0.51 GPN1GPN-loop GTPase 1 (ENSG00000198522), score: 0.72 GTF2E1general transcription factor IIE, polypeptide 1, alpha 56kDa (ENSG00000153767), score: 0.57 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.54 HM13histocompatibility (minor) 13 (ENSG00000101294), score: 0.54 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.51 HNF4Ghepatocyte nuclear factor 4, gamma (ENSG00000164749), score: 0.65 HTThuntingtin (ENSG00000197386), score: -0.74 HYIhydroxypyruvate isomerase (putative) (ENSG00000178922), score: 0.52 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (ENSG00000005700), score: 0.62 IL5RAinterleukin 5 receptor, alpha (ENSG00000091181), score: 0.52 IQCB1IQ motif containing B1 (ENSG00000173226), score: -0.62 ITCHitchy E3 ubiquitin protein ligase homolog (mouse) (ENSG00000078747), score: 0.53 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (ENSG00000115232), score: 0.68 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.61 KCTD6potassium channel tetramerisation domain containing 6 (ENSG00000168301), score: 0.54 KDM1Blysine (K)-specific demethylase 1B (ENSG00000165097), score: 0.55 KIAA0090KIAA0090 (ENSG00000127463), score: 0.56 KIAA0226KIAA0226 (ENSG00000145016), score: -0.56 KIAA0907KIAA0907 (ENSG00000132680), score: 0.67 KIAA1407KIAA1407 (ENSG00000163617), score: 0.53 KLF12Kruppel-like factor 12 (ENSG00000118922), score: -0.6 KLHDC8Bkelch domain containing 8B (ENSG00000185909), score: -0.6 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.57 KLHL8kelch-like 8 (Drosophila) (ENSG00000145332), score: 0.7 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (ENSG00000117009), score: 0.59 LACTBlactamase, beta (ENSG00000103642), score: 0.54 LAMB3laminin, beta 3 (ENSG00000196878), score: 0.56 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.66 LEAP2liver expressed antimicrobial peptide 2 (ENSG00000164406), score: 0.58 LIPClipase, hepatic (ENSG00000166035), score: 0.61 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: -0.63 LOC100302652GPR75-ASB3 (ENSG00000115239), score: 0.6 LRIG3leucine-rich repeats and immunoglobulin-like domains 3 (ENSG00000139263), score: 0.7 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.7 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.84 LRRC58leucine rich repeat containing 58 (ENSG00000163428), score: 0.61 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.58 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: 0.56 MAP2K5mitogen-activated protein kinase kinase 5 (ENSG00000137764), score: -0.75 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: -0.57 MARK3MAP/microtubule affinity-regulating kinase 3 (ENSG00000075413), score: -0.57 MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.57 MBNL2muscleblind-like 2 (Drosophila) (ENSG00000139793), score: -0.8 MCM6minichromosome maintenance complex component 6 (ENSG00000076003), score: 0.51 MED23mediator complex subunit 23 (ENSG00000112282), score: 0.66 MKLN1muskelin 1, intracellular mediator containing kelch motifs (ENSG00000128585), score: 0.54 MLLT1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 (ENSG00000130382), score: -0.56 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.58 MPDZmultiple PDZ domain protein (ENSG00000107186), score: -0.63 MRPL12mitochondrial ribosomal protein L12 (ENSG00000183093), score: -0.66 MSRAmethionine sulfoxide reductase A (ENSG00000175806), score: 0.55 MTA1metastasis associated 1 (ENSG00000182979), score: -0.71 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: 0.66 MYNNmyoneurin (ENSG00000085274), score: 0.69 N6AMT2N-6 adenine-specific DNA methyltransferase 2 (putative) (ENSG00000150456), score: 0.54 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.59 NCSTNnicastrin (ENSG00000162736), score: 0.6 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: 0.62 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.66 NRBP1nuclear receptor binding protein 1 (ENSG00000115216), score: -0.69 NT5DC25'-nucleotidase domain containing 2 (ENSG00000168268), score: -0.57 NT5E5'-nucleotidase, ecto (CD73) (ENSG00000135318), score: 0.81 NUP37nucleoporin 37kDa (ENSG00000075188), score: 0.52 OLFML2Bolfactomedin-like 2B (ENSG00000162745), score: 0.54 OSTM1osteopetrosis associated transmembrane protein 1 (ENSG00000081087), score: 0.59 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.68 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.54 PARP6poly (ADP-ribose) polymerase family, member 6 (ENSG00000137817), score: -0.75 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.53 PDRG1p53 and DNA-damage regulated 1 (ENSG00000088356), score: 0.53 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.69 PGM3phosphoglucomutase 3 (ENSG00000013375), score: 0.88 PHF16PHD finger protein 16 (ENSG00000102221), score: 0.52 PHOSPHO1phosphatase, orphan 1 (ENSG00000173868), score: 0.52 PIAS1protein inhibitor of activated STAT, 1 (ENSG00000033800), score: -0.6 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.62 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (ENSG00000186111), score: -0.59 PLDNpallidin homolog (mouse) (ENSG00000104164), score: 0.55 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (ENSG00000175895), score: 0.54 POFUT1protein O-fucosyltransferase 1 (ENSG00000101346), score: 0.52 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.63 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta (ENSG00000112640), score: -0.56 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: 0.51 QPCTglutaminyl-peptide cyclotransferase (ENSG00000115828), score: 0.71 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.73 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: 0.56 RANBP3RAN binding protein 3 (ENSG00000031823), score: -0.71 RECKreversion-inducing-cysteine-rich protein with kazal motifs (ENSG00000122707), score: 0.71 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: 0.56 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.52 RPAINRPA interacting protein (ENSG00000129197), score: 0.59 SCUBE3signal peptide, CUB domain, EGF-like 3 (ENSG00000146197), score: 0.62 SERINC5serine incorporator 5 (ENSG00000164300), score: 0.66 SERPINB5serpin peptidase inhibitor, clade B (ovalbumin), member 5 (ENSG00000206075), score: 0.58 SETD5SET domain containing 5 (ENSG00000168137), score: -0.77 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: -0.59 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (ENSG00000204351), score: 0.59 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (ENSG00000119899), score: 0.73 SLC30A1solute carrier family 30 (zinc transporter), member 1 (ENSG00000170385), score: 0.53 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.58 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.58 SLC31A1solute carrier family 31 (copper transporters), member 1 (ENSG00000136868), score: 0.57 SLC39A9solute carrier family 39 (zinc transporter), member 9 (ENSG00000029364), score: 0.93 SLC40A1solute carrier family 40 (iron-regulated transporter), member 1 (ENSG00000138449), score: 0.52 SLC45A3solute carrier family 45, member 3 (ENSG00000158715), score: 0.59 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.51 SMYD4SET and MYND domain containing 4 (ENSG00000186532), score: -0.59 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: -0.66 SNRNP48small nuclear ribonucleoprotein 48kDa (U11/U12) (ENSG00000168566), score: 0.54 SNX24sorting nexin 24 (ENSG00000064652), score: 0.52 SPATA5L1spermatogenesis associated 5-like 1 (ENSG00000171763), score: 0.56 SPENspen homolog, transcriptional regulator (Drosophila) (ENSG00000065526), score: -0.64 SPTLC2serine palmitoyltransferase, long chain base subunit 2 (ENSG00000100596), score: 0.53 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.65 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: -0.72 SS18synovial sarcoma translocation, chromosome 18 (ENSG00000141380), score: 0.58 ST8SIA4ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (ENSG00000113532), score: 0.51 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.72 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.53 TAAR1trace amine associated receptor 1 (ENSG00000146399), score: 0.53 TARSL2threonyl-tRNA synthetase-like 2 (ENSG00000185418), score: -0.72 TCF12transcription factor 12 (ENSG00000140262), score: 0.66 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.63 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.62 TMCC1transmembrane and coiled-coil domain family 1 (ENSG00000172765), score: -0.59 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.56 TMEM27transmembrane protein 27 (ENSG00000147003), score: 0.53 TMEM33transmembrane protein 33 (ENSG00000109133), score: 0.67 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.6 TPST1tyrosylprotein sulfotransferase 1 (ENSG00000169902), score: 0.58 TRIP13thyroid hormone receptor interactor 13 (ENSG00000071539), score: 0.6 TSHRthyroid stimulating hormone receptor (ENSG00000165409), score: 0.56 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.64 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: -0.63 TTC13tetratricopeptide repeat domain 13 (ENSG00000143643), score: 0.56 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.71 TWISTNBTWIST neighbor (ENSG00000105849), score: 0.66 TXNDC15thioredoxin domain containing 15 (ENSG00000113621), score: 0.62 UBA6ubiquitin-like modifier activating enzyme 6 (ENSG00000033178), score: 0.67 UBE2CBPubiquitin-conjugating enzyme E2C binding protein (ENSG00000118420), score: 0.55 UBL3ubiquitin-like 3 (ENSG00000122042), score: 0.69 UBXN4UBX domain protein 4 (ENSG00000144224), score: 0.58 UHRF1BP1LUHRF1 binding protein 1-like (ENSG00000111647), score: 0.59 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: 0.63 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: -0.61 USP20ubiquitin specific peptidase 20 (ENSG00000136878), score: -0.78 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: 0.6 VAT1vesicle amine transport protein 1 homolog (T. californica) (ENSG00000108828), score: 0.66 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.71 VPS4Bvacuolar protein sorting 4 homolog B (S. cerevisiae) (ENSG00000119541), score: 0.66 WDR3WD repeat domain 3 (ENSG00000065183), score: 0.63 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.63 WDR76WD repeat domain 76 (ENSG00000092470), score: 0.56 WDR8WD repeat domain 8 (ENSG00000116213), score: -0.62 WNT9Awingless-type MMTV integration site family, member 9A (ENSG00000143816), score: 0.55 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 0.78 XRCC6BP1XRCC6 binding protein 1 (ENSG00000166896), score: 0.66 YIPF5Yip1 domain family, member 5 (ENSG00000145817), score: 0.57 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.72 ZBTB49zinc finger and BTB domain containing 49 (ENSG00000168826), score: -0.72 ZBTB8Azinc finger and BTB domain containing 8A (ENSG00000160062), score: 0.53 ZC3H12Dzinc finger CCCH-type containing 12D (ENSG00000178199), score: 1 ZHX1zinc fingers and homeoboxes 1 (ENSG00000165156), score: 0.56 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (ENSG00000084073), score: 0.62 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.75 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.74 ZW10ZW10, kinetochore associated, homolog (Drosophila) (ENSG00000086827), score: 0.66
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_kd_m_ca1 | oan | kd | m | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |