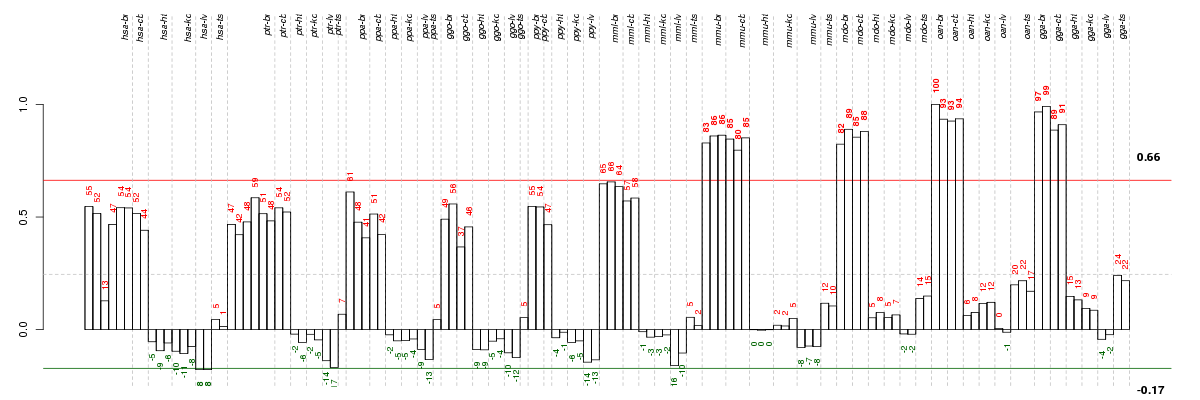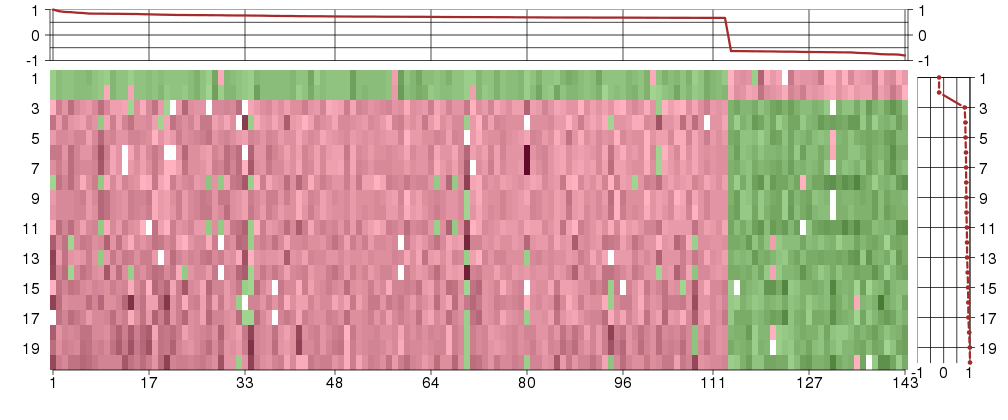

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
ion transport
The directed movement of charged atoms or small charged molecules into, out of, within or between cells by means of some external agent such as a transporter or pore.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signaling pathway
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
G-protein coupled receptor protein signaling pathway
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand.
gamma-aminobutyric acid signaling pathway
The series of molecular signals generated by the binding of gamma-aminobutyric acid (GABA, 4-aminobutyrate), an amino acid which acts as a neurotransmitter in some organisms, to a cell surface receptor.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
nerve-nerve synaptic transmission
The process of communication from a neuron to another neuron across a synapse.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
synaptic transmission, glutamatergic
The process of communication from a neuron to another neuron across a synapse using the neurotransmitter glutamate.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
all
NA
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
extracellular ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific extracellular ligand has been bound by the channel complex or one of its constituent parts.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
calcium channel activity
Catalysis of facilitated diffusion of a calcium ion (by an energy-independent process) involving passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
store-operated calcium channel activity
A ligand-gated ion channel activity which transports calcium in response to emptying of intracellular calcium stores.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
ligand-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
ligand-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a channel that opens when a specific ligand has been bound by the channel complex or one of its constituent parts.
store-operated calcium channel activity
A ligand-gated ion channel activity which transports calcium in response to emptying of intracellular calcium stores.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 8.708e-03 | 2.584 | 10 | 102 | Neuroactive ligand-receptor interaction |
| 00601 | 2.263e-02 | 0.1773 | 3 | 7 | Glycosphingolipid biosynthesis - lacto and neolacto series |
ABHD3abhydrolase domain containing 3 (ENSG00000158201), score: 0.72 ADAM22ADAM metallopeptidase domain 22 (ENSG00000008277), score: 0.74 ADCYAP1adenylate cyclase activating polypeptide 1 (pituitary) (ENSG00000141433), score: 0.69 ANO3anoctamin 3 (ENSG00000134343), score: 0.78 ARPP21cAMP-regulated phosphoprotein, 21kDa (ENSG00000172995), score: 0.73 ARRDC4arrestin domain containing 4 (ENSG00000140450), score: -0.67 ASAMadipocyte-specific adhesion molecule (ENSG00000166250), score: 0.71 ATP13A5ATPase type 13A5 (ENSG00000187527), score: 0.68 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (ENSG00000113732), score: -0.66 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.84 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (ENSG00000086062), score: -0.68 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (ENSG00000118276), score: 0.71 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.68 C14orf159chromosome 14 open reading frame 159 (ENSG00000133943), score: -0.63 C14orf37chromosome 14 open reading frame 37 (ENSG00000139971), score: 0.7 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.81 C6orf72chromosome 6 open reading frame 72 (ENSG00000055211), score: -0.68 C9orf4chromosome 9 open reading frame 4 (ENSG00000136805), score: 0.72 CACHD1cache domain containing 1 (ENSG00000158966), score: 0.68 CACNB4calcium channel, voltage-dependent, beta 4 subunit (ENSG00000182389), score: 0.72 CACNG2calcium channel, voltage-dependent, gamma subunit 2 (ENSG00000166862), score: 0.75 CASD1CAS1 domain containing 1 (ENSG00000127995), score: 0.68 CASP7caspase 7, apoptosis-related cysteine peptidase (ENSG00000165806), score: -0.67 CASTcalpastatin (ENSG00000153113), score: -0.71 CCBL2cysteine conjugate-beta lyase 2 (ENSG00000137944), score: -0.64 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.72 CDK17cyclin-dependent kinase 17 (ENSG00000059758), score: 0.83 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.87 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: -0.73 CLCN3chloride channel 3 (ENSG00000109572), score: 0.69 CLVS1clavesin 1 (ENSG00000177182), score: 0.71 CLVS2clavesin 2 (ENSG00000146352), score: 0.73 CNKSR2connector enhancer of kinase suppressor of Ras 2 (ENSG00000149970), score: 0.7 CNR1cannabinoid receptor 1 (brain) (ENSG00000118432), score: 0.76 CNTN4contactin 4 (ENSG00000144619), score: 0.69 CNTN5contactin 5 (ENSG00000149972), score: 0.79 COL25A1collagen, type XXV, alpha 1 (ENSG00000188517), score: 0.78 CRTAPcartilage associated protein (ENSG00000170275), score: -0.7 CSMD2CUB and Sushi multiple domains 2 (ENSG00000121904), score: 0.75 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: -0.75 CTTNBP2cortactin binding protein 2 (ENSG00000077063), score: 0.67 DCTDdCMP deaminase (ENSG00000129187), score: -0.63 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.86 DNAJC27DnaJ (Hsp40) homolog, subfamily C, member 27 (ENSG00000115137), score: 0.68 ELMOD1ELMO/CED-12 domain containing 1 (ENSG00000110675), score: 0.7 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (ENSG00000118402), score: 0.68 EN1engrailed homeobox 1 (ENSG00000163064), score: 0.7 EPHA3EPH receptor A3 (ENSG00000044524), score: 0.74 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.81 EPHA8EPH receptor A8 (ENSG00000070886), score: 0.78 EPHX4epoxide hydrolase 4 (ENSG00000172031), score: 0.78 FADDFas (TNFRSF6)-associated via death domain (ENSG00000168040), score: -0.76 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.71 FRMPD4FERM and PDZ domain containing 4 (ENSG00000169933), score: 0.72 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.69 GABRA2gamma-aminobutyric acid (GABA) A receptor, alpha 2 (ENSG00000151834), score: 0.7 GABRA4gamma-aminobutyric acid (GABA) A receptor, alpha 4 (ENSG00000109158), score: 0.71 GABRB2gamma-aminobutyric acid (GABA) A receptor, beta 2 (ENSG00000145864), score: 0.67 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.82 GALNTL6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 (ENSG00000174473), score: 0.68 GLRA1glycine receptor, alpha 1 (ENSG00000145888), score: 0.69 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.73 GPM6Bglycoprotein M6B (ENSG00000046653), score: 0.67 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 1 GPR37L1G protein-coupled receptor 37 like 1 (ENSG00000170075), score: 0.68 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.67 GRIA1glutamate receptor, ionotropic, AMPA 1 (ENSG00000155511), score: 0.7 GRIA4glutamate receptor, ionotrophic, AMPA 4 (ENSG00000152578), score: 0.69 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.8 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.74 IAH1isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) (ENSG00000134330), score: -0.64 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.83 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.7 KCNA1potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) (ENSG00000111262), score: 0.7 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.9 KIAA0391KIAA0391 (ENSG00000100890), score: -0.76 KIAA1279KIAA1279 (ENSG00000198954), score: 0.72 KIAA1467KIAA1467 (ENSG00000084444), score: 0.69 KIAA2022KIAA2022 (ENSG00000050030), score: 0.79 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.93 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.8 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.73 LPPR4lipid phosphate phosphatase-related protein type 4 (ENSG00000117600), score: 0.69 LRFN5leucine rich repeat and fibronectin type III domain containing 5 (ENSG00000165379), score: 0.68 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.67 LRRN1leucine rich repeat neuronal 1 (ENSG00000175928), score: 0.77 LSM10LSM10, U7 small nuclear RNA associated (ENSG00000181817), score: -0.72 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.64 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.83 MMP16matrix metallopeptidase 16 (membrane-inserted) (ENSG00000156103), score: 0.75 MRPL21mitochondrial ribosomal protein L21 (ENSG00000197345), score: -0.63 MYO16myosin XVI (ENSG00000041515), score: 0.83 NKAIN4Na+/K+ transporting ATPase interacting 4 (ENSG00000101198), score: 0.69 NPY5Rneuropeptide Y receptor Y5 (ENSG00000164129), score: 0.68 NTRK3neurotrophic tyrosine kinase, receptor, type 3 (ENSG00000140538), score: 0.71 NTSneurotensin (ENSG00000133636), score: 0.82 OSTF1osteoclast stimulating factor 1 (ENSG00000134996), score: -0.67 PAFAH1B2platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 (30kDa) (ENSG00000168092), score: 0.67 PCSK2proprotein convertase subtilisin/kexin type 2 (ENSG00000125851), score: 0.71 PDE10Aphosphodiesterase 10A (ENSG00000112541), score: 0.69 PEX5Lperoxisomal biogenesis factor 5-like (ENSG00000114757), score: 0.7 PPP3CAprotein phosphatase 3, catalytic subunit, alpha isozyme (ENSG00000138814), score: 0.73 PQLC1PQ loop repeat containing 1 (ENSG00000122490), score: -0.65 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.77 PTP4A2protein tyrosine phosphatase type IVA, member 2 (ENSG00000184007), score: -0.65 PTPRRprotein tyrosine phosphatase, receptor type, R (ENSG00000153233), score: 0.73 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: -0.66 RAB3GAP2RAB3 GTPase activating protein subunit 2 (non-catalytic) (ENSG00000118873), score: 0.76 RGS7BPregulator of G-protein signaling 7 binding protein (ENSG00000186479), score: 0.78 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.9 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (ENSG00000137275), score: -0.64 SCAMP1secretory carrier membrane protein 1 (ENSG00000085365), score: 0.73 SCG2secretogranin II (ENSG00000171951), score: 0.72 SEMA3Asema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A (ENSG00000075213), score: 0.81 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.77 SGTBsmall glutamine-rich tetratricopeptide repeat (TPR)-containing, beta (ENSG00000197860), score: 0.7 SLC12A5solute carrier family 12 (potassium/chloride transporter), member 5 (ENSG00000124140), score: 0.67 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.84 SLC4A10solute carrier family 4, sodium bicarbonate transporter, member 10 (ENSG00000144290), score: 0.69 SLC5A7solute carrier family 5 (choline transporter), member 7 (ENSG00000115665), score: 0.76 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (ENSG00000132164), score: 0.75 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.68 SLC7A14solute carrier family 7 (cationic amino acid transporter, y+ system), member 14 (ENSG00000013293), score: 0.68 SNAP23synaptosomal-associated protein, 23kDa (ENSG00000092531), score: -0.67 SNX25sorting nexin 25 (ENSG00000109762), score: 0.68 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (ENSG00000090487), score: -0.64 SPHKAPSPHK1 interactor, AKAP domain containing (ENSG00000153820), score: 0.72 SPON1spondin 1, extracellular matrix protein (ENSG00000152268), score: 0.71 STX12syntaxin 12 (ENSG00000117758), score: 0.68 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.69 SYNJ1synaptojanin 1 (ENSG00000159082), score: 0.78 TBC1D30TBC1 domain family, member 30 (ENSG00000111490), score: 0.67 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (ENSG00000198959), score: -0.68 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.84 TMEFF2transmembrane protein with EGF-like and two follistatin-like domains 2 (ENSG00000144339), score: 0.69 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (ENSG00000185215), score: -0.65 TRPC1transient receptor potential cation channel, subfamily C, member 1 (ENSG00000144935), score: 0.72 TRPC3transient receptor potential cation channel, subfamily C, member 3 (ENSG00000138741), score: 0.77 TRPC5transient receptor potential cation channel, subfamily C, member 5 (ENSG00000072315), score: 0.77 TSPAN2tetraspanin 2 (ENSG00000134198), score: 0.77 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: 0.68 WDFY3WD repeat and FYVE domain containing 3 (ENSG00000163625), score: 0.78
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| mmu_cb_m2_ca1 | mmu | cb | m | 2 |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mmu_br_m2_ca1 | mmu | br | m | 2 |
| mmu_cb_m1_ca1 | mmu | cb | m | 1 |
| mmu_cb_f_ca1 | mmu | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mmu_br_m1_ca1 | mmu | br | m | 1 |
| mmu_br_f_ca1 | mmu | br | f | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| gga_cb_m_ca1 | gga | cb | m | _ |
| mdo_br_f_ca1 | mdo | br | f | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| gga_br_m_ca1 | gga | br | m | _ |
| gga_br_f_ca1 | gga | br | f | _ |
| oan_br_m_ca1 | oan | br | m | _ |