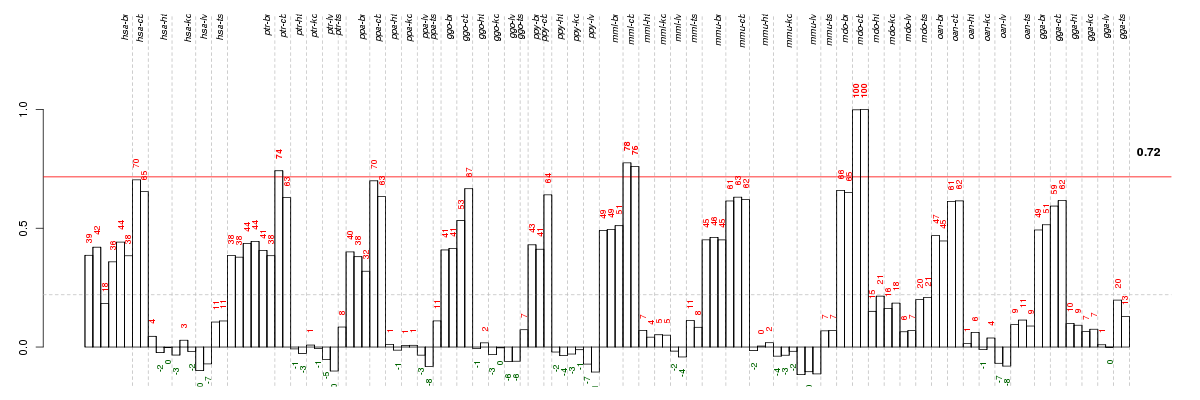

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
NA
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.

cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
neuron projection
A prolongation or process extending from a nerve cell, e.g. an axon or dendrite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

ABCC8ATP-binding cassette, sub-family C (CFTR/MRP), member 8 (ENSG00000006071), score: 0.46 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (ENSG00000172350), score: 0.48 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: 0.49 ADAM22ADAM metallopeptidase domain 22 (ENSG00000008277), score: 0.5 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.52 AIMP2aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 (ENSG00000106305), score: -0.51 AKAP13A kinase (PRKA) anchor protein 13 (ENSG00000170776), score: -0.49 AKT3v-akt murine thymoma viral oncogene homolog 3 (protein kinase B, gamma) (ENSG00000117020), score: 0.44 ALG14asparagine-linked glycosylation 14 homolog (S. cerevisiae) (ENSG00000172339), score: -0.61 ALKBH2alkB, alkylation repair homolog 2 (E. coli) (ENSG00000189046), score: 0.54 AP3B2adaptor-related protein complex 3, beta 2 subunit (ENSG00000103723), score: 0.43 APPL2adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 (ENSG00000136044), score: 0.52 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: -0.48 ARHGAP26Rho GTPase activating protein 26 (ENSG00000145819), score: 0.47 ARHGDIGRho GDP dissociation inhibitor (GDI) gamma (ENSG00000167930), score: -0.62 ARID1BAT rich interactive domain 1B (SWI1-like) (ENSG00000049618), score: 0.52 ARRDC4arrestin domain containing 4 (ENSG00000140450), score: -0.5 ASAMadipocyte-specific adhesion molecule (ENSG00000166250), score: 0.68 ASB1ankyrin repeat and SOCS box-containing 1 (ENSG00000065802), score: 0.45 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (ENSG00000113732), score: -0.58 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (ENSG00000086062), score: -0.51 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (ENSG00000118276), score: 0.52 BCAR3breast cancer anti-estrogen resistance 3 (ENSG00000137936), score: -0.48 BCDIN3DBCDIN3 domain containing (ENSG00000186666), score: 0.48 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: -0.53 BCL7AB-cell CLL/lymphoma 7A (ENSG00000110987), score: 0.47 BCL9B-cell CLL/lymphoma 9 (ENSG00000116128), score: 0.53 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.58 BRPF1bromodomain and PHD finger containing, 1 (ENSG00000156983), score: 0.45 BTBD11BTB (POZ) domain containing 11 (ENSG00000151136), score: 0.52 BTBD3BTB (POZ) domain containing 3 (ENSG00000132640), score: 0.48 C11orf41chromosome 11 open reading frame 41 (ENSG00000110427), score: 0.49 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.44 C11orf61chromosome 11 open reading frame 61 (ENSG00000120458), score: 0.49 C12orf53chromosome 12 open reading frame 53 (ENSG00000139200), score: 0.45 C13orf23chromosome 13 open reading frame 23 (ENSG00000120685), score: 0.71 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.62 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: 0.48 C17orf75chromosome 17 open reading frame 75 (ENSG00000108666), score: 0.43 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.5 C3orf18chromosome 3 open reading frame 18 (ENSG00000088543), score: 0.44 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.55 C3orf58chromosome 3 open reading frame 58 (ENSG00000181744), score: 0.48 C3orf59chromosome 3 open reading frame 59 (ENSG00000180611), score: 0.6 C6orf115chromosome 6 open reading frame 115 (ENSG00000146386), score: -0.59 C6orf145chromosome 6 open reading frame 145 (ENSG00000168994), score: -0.51 C6orf64chromosome 6 open reading frame 64 (ENSG00000112167), score: -0.65 C6orf72chromosome 6 open reading frame 72 (ENSG00000055211), score: -0.58 C8orf79chromosome 8 open reading frame 79 (ENSG00000170941), score: 0.69 CA10carbonic anhydrase X (ENSG00000154975), score: 0.47 CA8carbonic anhydrase VIII (ENSG00000178538), score: 0.76 CACNA1Gcalcium channel, voltage-dependent, T type, alpha 1G subunit (ENSG00000006283), score: 0.67 CACNG2calcium channel, voltage-dependent, gamma subunit 2 (ENSG00000166862), score: 0.48 CALB1calbindin 1, 28kDa (ENSG00000104327), score: 0.5 CAMKK2calcium/calmodulin-dependent protein kinase kinase 2, beta (ENSG00000110931), score: 0.55 CASP2caspase 2, apoptosis-related cysteine peptidase (ENSG00000106144), score: 0.43 CASP7caspase 7, apoptosis-related cysteine peptidase (ENSG00000165806), score: -0.56 CASP9caspase 9, apoptosis-related cysteine peptidase (ENSG00000132906), score: 0.58 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (ENSG00000078699), score: 0.58 CBFA2T3core-binding factor, runt domain, alpha subunit 2; translocated to, 3 (ENSG00000129993), score: 0.44 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.74 CCNT2cyclin T2 (ENSG00000082258), score: 0.67 CDH18cadherin 18, type 2 (ENSG00000145526), score: 0.44 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.73 CDR2Lcerebellar degeneration-related protein 2-like (ENSG00000109089), score: 0.48 CHGBchromogranin B (secretogranin 1) (ENSG00000089199), score: 0.51 CHN2chimerin (chimaerin) 2 (ENSG00000106069), score: 0.58 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: -0.49 CLVS2clavesin 2 (ENSG00000146352), score: 0.53 CNKSR2connector enhancer of kinase suppressor of Ras 2 (ENSG00000149970), score: 0.47 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: 0.56 CNR1cannabinoid receptor 1 (brain) (ENSG00000118432), score: 0.61 CNTN4contactin 4 (ENSG00000144619), score: 0.52 CNTN6contactin 6 (ENSG00000134115), score: 0.51 CNTNAP1contactin associated protein 1 (ENSG00000108797), score: 0.48 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.62 CPA6carboxypeptidase A6 (ENSG00000165078), score: 0.44 CRB1crumbs homolog 1 (Drosophila) (ENSG00000134376), score: 0.48 CRCPCGRP receptor component (ENSG00000241258), score: 0.48 CRHR1corticotropin releasing hormone receptor 1 (ENSG00000120088), score: 0.55 CRISPLD2cysteine-rich secretory protein LCCL domain containing 2 (ENSG00000103196), score: -0.48 CRTAPcartilage associated protein (ENSG00000170275), score: -0.63 CTCFCCCTC-binding factor (zinc finger protein) (ENSG00000102974), score: 0.46 CTNNB1catenin (cadherin-associated protein), beta 1, 88kDa (ENSG00000168036), score: 0.5 CTNNBIP1catenin, beta interacting protein 1 (ENSG00000178585), score: 0.44 CTSHcathepsin H (ENSG00000103811), score: -0.53 CYR61cysteine-rich, angiogenic inducer, 61 (ENSG00000142871), score: -0.48 CYTBcytochrome b (ENSG00000198727), score: -0.48 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (ENSG00000100201), score: 0.43 DDX47DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (ENSG00000213782), score: -0.49 DGKEdiacylglycerol kinase, epsilon 64kDa (ENSG00000153933), score: 0.43 DGKZdiacylglycerol kinase, zeta 104kDa (ENSG00000149091), score: 0.44 DHRS13dehydrogenase/reductase (SDR family) member 13 (ENSG00000167536), score: 0.47 DHRS7Bdehydrogenase/reductase (SDR family) member 7B (ENSG00000109016), score: -0.49 DNAJC1DnaJ (Hsp40) homolog, subfamily C, member 1 (ENSG00000136770), score: -0.49 DNERdelta/notch-like EGF repeat containing (ENSG00000187957), score: 0.43 DNM3dynamin 3 (ENSG00000197959), score: 0.51 DPP6dipeptidyl-peptidase 6 (ENSG00000130226), score: 0.47 DSEdermatan sulfate epimerase (ENSG00000111817), score: -0.48 E4F1E4F transcription factor 1 (ENSG00000167967), score: 0.53 EBF1early B-cell factor 1 (ENSG00000164330), score: 0.46 ECE2endothelin converting enzyme 2 (ENSG00000145194), score: 0.49 EDC4enhancer of mRNA decapping 4 (ENSG00000038358), score: 0.51 EIF3Beukaryotic translation initiation factor 3, subunit B (ENSG00000106263), score: -0.54 EIF3Ieukaryotic translation initiation factor 3, subunit I (ENSG00000084623), score: -0.58 EN1engrailed homeobox 1 (ENSG00000163064), score: 0.59 EPC1enhancer of polycomb homolog 1 (Drosophila) (ENSG00000120616), score: 0.57 EPGNepithelial mitogen homolog (mouse) (ENSG00000182585), score: 0.82 EPHA8EPH receptor A8 (ENSG00000070886), score: 0.62 ERBB2IPerbb2 interacting protein (ENSG00000112851), score: -0.58 ERGv-ets erythroblastosis virus E26 oncogene homolog (avian) (ENSG00000157554), score: -0.49 FAM116Bfamily with sequence similarity 116, member B (ENSG00000205593), score: 0.43 FAM131Bfamily with sequence similarity 131, member B (ENSG00000159784), score: 0.46 FAM135Bfamily with sequence similarity 135, member B (ENSG00000147724), score: 0.44 FAM13Cfamily with sequence similarity 13, member C (ENSG00000148541), score: 0.47 FAM73Afamily with sequence similarity 73, member A (ENSG00000180488), score: 0.53 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.87 FGF14fibroblast growth factor 14 (ENSG00000102466), score: 0.51 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.91 FGF9fibroblast growth factor 9 (glia-activating factor) (ENSG00000102678), score: 0.45 FGFR1fibroblast growth factor receptor 1 (ENSG00000077782), score: 0.48 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.44 FKBP5FK506 binding protein 5 (ENSG00000096060), score: -0.73 FKBP9FK506 binding protein 9, 63 kDa (ENSG00000122642), score: -0.51 FNDC5fibronectin type III domain containing 5 (ENSG00000160097), score: 0.43 FOXO3forkhead box O3 (ENSG00000118689), score: 0.46 FUBP1far upstream element (FUSE) binding protein 1 (ENSG00000162613), score: 0.55 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.5 GABRB2gamma-aminobutyric acid (GABA) A receptor, beta 2 (ENSG00000145864), score: 0.47 GABRG2gamma-aminobutyric acid (GABA) A receptor, gamma 2 (ENSG00000113327), score: 0.43 GALNT7UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) (ENSG00000109586), score: 0.6 GAR1GAR1 ribonucleoprotein homolog (yeast) (ENSG00000109534), score: -0.5 GBF1golgi brefeldin A resistant guanine nucleotide exchange factor 1 (ENSG00000107862), score: 0.48 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (ENSG00000158555), score: 0.43 GEMIN6gem (nuclear organelle) associated protein 6 (ENSG00000152147), score: -0.5 GHITMgrowth hormone inducible transmembrane protein (ENSG00000165678), score: -0.65 GIT2G protein-coupled receptor kinase interacting ArfGAP 2 (ENSG00000139436), score: 0.51 GLCEglucuronic acid epimerase (ENSG00000138604), score: 0.53 GNG13guanine nucleotide binding protein (G protein), gamma 13 (ENSG00000127588), score: 0.57 GPR37L1G protein-coupled receptor 37 like 1 (ENSG00000170075), score: 0.46 GRB14growth factor receptor-bound protein 14 (ENSG00000115290), score: -0.5 GRIA4glutamate receptor, ionotrophic, AMPA 4 (ENSG00000152578), score: 0.53 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.74 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.61 H2AFY2H2A histone family, member Y2 (ENSG00000099284), score: 0.58 HADHhydroxyacyl-CoA dehydrogenase (ENSG00000138796), score: -0.53 HCN1hyperpolarization activated cyclic nucleotide-gated potassium channel 1 (ENSG00000164588), score: 0.45 HMG20Ahigh-mobility group 20A (ENSG00000140382), score: 0.54 ICMTisoprenylcysteine carboxyl methyltransferase (ENSG00000116237), score: 0.45 IGDCC3immunoglobulin superfamily, DCC subclass, member 3 (ENSG00000174498), score: 0.43 IGDCC4immunoglobulin superfamily, DCC subclass, member 4 (ENSG00000103742), score: 0.46 IL10RBinterleukin 10 receptor, beta (ENSG00000243646), score: -0.48 IL17RAinterleukin 17 receptor A (ENSG00000177663), score: -0.65 INVSinversin (ENSG00000119509), score: -0.63 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.49 KCNA1potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) (ENSG00000111262), score: 0.51 KCNC1potassium voltage-gated channel, Shaw-related subfamily, member 1 (ENSG00000129159), score: 0.57 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (ENSG00000171385), score: 0.47 KCNIP1Kv channel interacting protein 1 (ENSG00000182132), score: 0.57 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.51 KCNK10potassium channel, subfamily K, member 10 (ENSG00000100433), score: 0.71 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.52 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.45 KCTD6potassium channel tetramerisation domain containing 6 (ENSG00000168301), score: 0.65 KDM4Alysine (K)-specific demethylase 4A (ENSG00000066135), score: 0.44 KIAA0240KIAA0240 (ENSG00000112624), score: 0.48 KIAA0895KIAA0895 (ENSG00000164542), score: 0.43 KIAA1217KIAA1217 (ENSG00000120549), score: -0.63 KIAA1409KIAA1409 (ENSG00000133958), score: 0.45 KIAA2022KIAA2022 (ENSG00000050030), score: 0.43 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: 0.46 LAMB1laminin, beta 1 (ENSG00000091136), score: -0.54 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.65 LOC100131509similar to inward rectifying K+ channel negative regulator Kir2.2v (ENSG00000184185), score: 0.47 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.43 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: 0.51 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (ENSG00000157193), score: 0.51 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.55 LRRC38leucine rich repeat containing 38 (ENSG00000162494), score: 0.6 LRRN1leucine rich repeat neuronal 1 (ENSG00000175928), score: 0.55 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: -0.57 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.82 MAML2mastermind-like 2 (Drosophila) (ENSG00000184384), score: 0.45 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.8 MAMLD1mastermind-like domain containing 1 (ENSG00000013619), score: 0.44 MAN2A1mannosidase, alpha, class 2A, member 1 (ENSG00000112893), score: -0.49 MCTP1multiple C2 domains, transmembrane 1 (ENSG00000175471), score: 0.46 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.52 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.57 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.49 MESDC1mesoderm development candidate 1 (ENSG00000140406), score: 0.45 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: -0.53 MID2midline 2 (ENSG00000080561), score: 0.55 MIXL1Mix1 homeobox-like 1 (Xenopus laevis) (ENSG00000185155), score: 1 MLLmyeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) (ENSG00000118058), score: 0.46 MMP16matrix metallopeptidase 16 (membrane-inserted) (ENSG00000156103), score: 0.56 MMRN2multimerin 2 (ENSG00000173269), score: -0.49 MRPL28mitochondrial ribosomal protein L28 (ENSG00000086504), score: -0.56 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.66 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.68 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.68 ND2MTND2 (ENSG00000198763), score: -0.5 ND4NADH dehydrogenase, subunit 4 (complex I) (ENSG00000198886), score: -0.5 NDE1nudE nuclear distribution gene E homolog 1 (A. nidulans) (ENSG00000072864), score: -0.53 NFIAnuclear factor I/A (ENSG00000162599), score: 0.44 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (ENSG00000100906), score: -0.48 NFKBIDnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta (ENSG00000167604), score: 0.52 NFRKBnuclear factor related to kappaB binding protein (ENSG00000170322), score: 0.44 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.43 NMNAT2nicotinamide nucleotide adenylyltransferase 2 (ENSG00000157064), score: 0.47 NR1H3nuclear receptor subfamily 1, group H, member 3 (ENSG00000025434), score: -0.5 NRG2neuregulin 2 (ENSG00000158458), score: 0.64 NRP1neuropilin 1 (ENSG00000099250), score: -0.48 NTF3neurotrophin 3 (ENSG00000185652), score: 0.55 NTMneurotrimin (ENSG00000182667), score: 0.48 NTN4netrin 4 (ENSG00000074527), score: -0.61 NTRK3neurotrophic tyrosine kinase, receptor, type 3 (ENSG00000140538), score: 0.48 NUB1negative regulator of ubiquitin-like proteins 1 (ENSG00000013374), score: -0.48 NUP85nucleoporin 85kDa (ENSG00000125450), score: 0.49 OGFOD12-oxoglutarate and iron-dependent oxygenase domain containing 1 (ENSG00000087263), score: -0.48 OLFM3olfactomedin 3 (ENSG00000118733), score: 0.52 ORAI2ORAI calcium release-activated calcium modulator 2 (ENSG00000160991), score: 0.53 OTOL1otolin 1 homolog (zebrafish) (ENSG00000182447), score: 0.66 P4HA2prolyl 4-hydroxylase, alpha polypeptide II (ENSG00000072682), score: -0.53 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (ENSG00000149269), score: 0.47 PARD3par-3 partitioning defective 3 homolog (C. elegans) (ENSG00000148498), score: -0.54 PAX3paired box 3 (ENSG00000135903), score: 0.6 PAX6paired box 6 (ENSG00000007372), score: 0.73 PDGFRLplatelet-derived growth factor receptor-like (ENSG00000104213), score: 0.52 PDZD7PDZ domain containing 7 (ENSG00000186862), score: 0.68 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.57 PHF21APHD finger protein 21A (ENSG00000135365), score: 0.56 PIGNphosphatidylinositol glycan anchor biosynthesis, class N (ENSG00000197563), score: -0.49 PIGSphosphatidylinositol glycan anchor biosynthesis, class S (ENSG00000087111), score: 0.57 PIK3R3phosphoinositide-3-kinase, regulatory subunit 3 (gamma) (ENSG00000117461), score: 0.49 PLCB4phospholipase C, beta 4 (ENSG00000101333), score: 0.6 PLCXD3phosphatidylinositol-specific phospholipase C, X domain containing 3 (ENSG00000182836), score: 0.47 PLD5phospholipase D family, member 5 (ENSG00000180287), score: 0.55 PLEKHF1pleckstrin homology domain containing, family F (with FYVE domain) member 1 (ENSG00000166289), score: -0.52 PLS3plastin 3 (ENSG00000102024), score: -0.48 PPA2pyrophosphatase (inorganic) 2 (ENSG00000138777), score: -0.53 PPARGperoxisome proliferator-activated receptor gamma (ENSG00000132170), score: -0.52 PPM1Hprotein phosphatase, Mg2+/Mn2+ dependent, 1H (ENSG00000111110), score: 0.49 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha (ENSG00000066027), score: -0.49 PPP3CAprotein phosphatase 3, catalytic subunit, alpha isozyme (ENSG00000138814), score: 0.43 PRDM11PR domain containing 11 (ENSG00000019485), score: 0.45 PRKCHprotein kinase C, eta (ENSG00000027075), score: -0.48 PRKD3protein kinase D3 (ENSG00000115825), score: -0.49 PSD2pleckstrin and Sec7 domain containing 2 (ENSG00000146005), score: 0.48 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.64 PTP4A2protein tyrosine phosphatase type IVA, member 2 (ENSG00000184007), score: -0.57 PTPN22protein tyrosine phosphatase, non-receptor type 22 (lymphoid) (ENSG00000134242), score: 0.63 PTPREprotein tyrosine phosphatase, receptor type, E (ENSG00000132334), score: 0.46 PTPRRprotein tyrosine phosphatase, receptor type, R (ENSG00000153233), score: 0.59 PVALBparvalbumin (ENSG00000100362), score: 0.55 RADILRas association and DIL domains (ENSG00000157927), score: 0.44 RAI14retinoic acid induced 14 (ENSG00000039560), score: -0.48 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.43 REXO2REX2, RNA exonuclease 2 homolog (S. cerevisiae) (ENSG00000076043), score: -0.53 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: -0.5 RGS7BPregulator of G-protein signaling 7 binding protein (ENSG00000186479), score: 0.54 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.76 RHBDF2rhomboid 5 homolog 2 (Drosophila) (ENSG00000129667), score: -0.56 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.52 RIN2Ras and Rab interactor 2 (ENSG00000132669), score: -0.5 RIPK2receptor-interacting serine-threonine kinase 2 (ENSG00000104312), score: -0.61 RNF122ring finger protein 122 (ENSG00000133874), score: 0.62 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (ENSG00000117676), score: 0.51 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.43 RUNX1T1runt-related transcription factor 1; translocated to, 1 (cyclin D-related) (ENSG00000079102), score: 0.46 RXRAretinoid X receptor, alpha (ENSG00000186350), score: -0.62 SBK1SH3-binding domain kinase 1 (ENSG00000188322), score: 0.44 SEC11ASEC11 homolog A (S. cerevisiae) (ENSG00000140612), score: -0.57 SEL1L3sel-1 suppressor of lin-12-like 3 (C. elegans) (ENSG00000091490), score: 0.56 SELSselenoprotein S (ENSG00000131871), score: -0.49 SENP7SUMO1/sentrin specific peptidase 7 (ENSG00000138468), score: 0.45 SETD5SET domain containing 5 (ENSG00000168137), score: 0.6 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.53 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.59 SGK3serum/glucocorticoid regulated kinase family, member 3 (ENSG00000104205), score: -0.57 SGSM1small G protein signaling modulator 1 (ENSG00000167037), score: 0.5 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: 0.46 SHFSrc homology 2 domain containing F (ENSG00000138606), score: 0.57 SHROOM2shroom family member 2 (ENSG00000146950), score: 0.48 SIDT1SID1 transmembrane family, member 1 (ENSG00000072858), score: 0.53 SIK1salt-inducible kinase 1 (ENSG00000142178), score: -0.5 SKAP2src kinase associated phosphoprotein 2 (ENSG00000005020), score: -0.52 SKIv-ski sarcoma viral oncogene homolog (avian) (ENSG00000157933), score: 0.48 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.83 SLC12A5solute carrier family 12 (potassium/chloride transporter), member 5 (ENSG00000124140), score: 0.45 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.5 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.74 SLC6A5solute carrier family 6 (neurotransmitter transporter, glycine), member 5 (ENSG00000165970), score: 0.68 SLC9A5solute carrier family 9 (sodium/hydrogen exchanger), member 5 (ENSG00000135740), score: 0.63 SLITRK4SLIT and NTRK-like family, member 4 (ENSG00000179542), score: 0.49 SMYD4SET and MYND domain containing 4 (ENSG00000186532), score: 0.52 SNAP23synaptosomal-associated protein, 23kDa (ENSG00000092531), score: -0.59 SNAP25synaptosomal-associated protein, 25kDa (ENSG00000132639), score: 0.47 SNAP91synaptosomal-associated protein, 91kDa homolog (mouse) (ENSG00000065609), score: 0.44 SNED1sushi, nidogen and EGF-like domains 1 (ENSG00000162804), score: 0.45 SNRKSNF related kinase (ENSG00000163788), score: 0.51 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (ENSG00000090487), score: -0.48 SPHKAPSPHK1 interactor, AKAP domain containing (ENSG00000153820), score: 0.59 SPTBspectrin, beta, erythrocytic (ENSG00000070182), score: 0.43 SRCIN1SRC kinase signaling inhibitor 1 (ENSG00000017373), score: 0.5 SRRM4serine/arginine repetitive matrix 4 (ENSG00000139767), score: 0.52 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.49 STX8syntaxin 8 (ENSG00000170310), score: -0.63 SUSD1sushi domain containing 1 (ENSG00000106868), score: -0.48 SYBUsyntabulin (syntaxin-interacting) (ENSG00000147642), score: 0.49 SYF2SYF2 homolog, RNA splicing factor (S. cerevisiae) (ENSG00000117614), score: -0.54 SYN2synapsin II (ENSG00000157152), score: 0.45 SYT12synaptotagmin XII (ENSG00000173227), score: 0.55 SYT4synaptotagmin IV (ENSG00000132872), score: 0.52 TBC1D1TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 (ENSG00000065882), score: -0.5 TFAP2Btranscription factor AP-2 beta (activating enhancer binding protein 2 beta) (ENSG00000008196), score: 0.44 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (ENSG00000198959), score: -0.63 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.44 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.57 TLL1tolloid-like 1 (ENSG00000038295), score: 0.71 TM4SF1transmembrane 4 L six family member 1 (ENSG00000169908), score: -0.48 TMEM120Btransmembrane protein 120B (ENSG00000188735), score: 0.54 TMEM63Ctransmembrane protein 63C (ENSG00000165548), score: 0.49 TMPRSS9transmembrane protease, serine 9 (ENSG00000178297), score: 0.46 TMTC2transmembrane and tetratricopeptide repeat containing 2 (ENSG00000179104), score: 0.43 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (ENSG00000185215), score: -0.56 TNRC6Atrinucleotide repeat containing 6A (ENSG00000090905), score: 0.48 TPH2tryptophan hydroxylase 2 (ENSG00000139287), score: 0.44 TRHDEthyrotropin-releasing hormone degrading enzyme (ENSG00000072657), score: 0.49 TRIM3tripartite motif-containing 3 (ENSG00000110171), score: 0.51 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.66 TRPC3transient receptor potential cation channel, subfamily C, member 3 (ENSG00000138741), score: 0.71 TUSC3tumor suppressor candidate 3 (ENSG00000104723), score: 0.53 TXNL1thioredoxin-like 1 (ENSG00000091164), score: -0.51 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (ENSG00000197355), score: -0.62 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.48 UGGT2UDP-glucose glycoprotein glucosyltransferase 2 (ENSG00000102595), score: -0.57 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: -0.51 UNC5Bunc-5 homolog B (C. elegans) (ENSG00000107731), score: 0.46 UNC80unc-80 homolog (C. elegans) (ENSG00000144406), score: 0.52 UPF0639UPF0639 protein (ENSG00000175985), score: 0.74 USP20ubiquitin specific peptidase 20 (ENSG00000136878), score: 0.53 USP3ubiquitin specific peptidase 3 (ENSG00000140455), score: 0.48 VAT1Lvesicle amine transport protein 1 homolog (T. californica)-like (ENSG00000171724), score: 0.43 VSNL1visinin-like 1 (ENSG00000163032), score: 0.49 WSCD1WSC domain containing 1 (ENSG00000179314), score: 0.55 ZBTB49zinc finger and BTB domain containing 49 (ENSG00000168826), score: 0.49 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.5 ZDHHC15zinc finger, DHHC-type containing 15 (ENSG00000102383), score: 0.43 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: 0.63 ZFYVE27zinc finger, FYVE domain containing 27 (ENSG00000155256), score: 0.54 ZIC4Zic family member 4 (ENSG00000174963), score: 0.87 ZMIZ1zinc finger, MIZ-type containing 1 (ENSG00000108175), score: 0.46 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.65 ZNF385Czinc finger protein 385C (ENSG00000187595), score: 0.65 ZNF462zinc finger protein 462 (ENSG00000148143), score: 0.53 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.47 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.67 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.48
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_cb_m_ca1 | ptr | cb | m | _ |
| mml_cb_f_ca1 | mml | cb | f | _ |
| mml_cb_m_ca1 | mml | cb | m | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |