|
Introduction
|
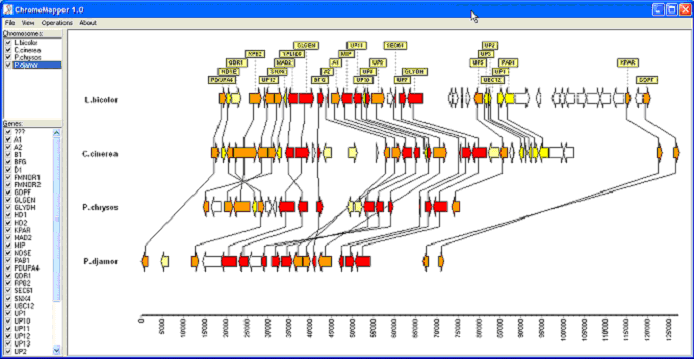
Overview ChromoMapper is a free, user-friendly tool designed to visualize the arrangement of genes in the genome and to emphasize syntenic relationships within or among genomes. Given several species with gene positions and consensus names, it assemble, display and quantify gene order relationships. Functionalities ChromoMapper represents the genes of each species (or chromosome) by labelled arrows indicating their relative transcriptional orientation. A grey or colour gradient indicates in how many species/chromosome each gene is conserved. Unnamed genes can be represented by empty arrows or omitted altogether. Optionally, connectors can join orthologue genes. ChromoMapper allows finding the chromosome orientations that best evidences the synteny. This is done by minimizing the Spearman correlation between gene ranks for all pairs of chromosomes (menu “Operations/Disentangle links”). Moreover, the orientation of each chromosome can be set manually. Simple mouse operations allow zooming on and exploring particular regions of the chromosomes.
The
display if largely customisable: e.g., it
is possible to switch on and off individual genomes and genes, or
orthologue
connectors, or to choose how gene names are displayed. Once the desired
gene
map is obtained, it can be exported as a WMF (Windows Metafile Format)
vector
image. Interactivity
is the major feature of
ChromoMapper. It allows sorting out overwhelmingly large amount of
information
to generate customizable, informative figures. |
|
Credits
|
Conceived by
Hélène Niculita-Hirzel & Alexandre H. Hirzel
Realised by Alexandre
H. Hirzel |
|
Bibliography
|
|
|
Download
|