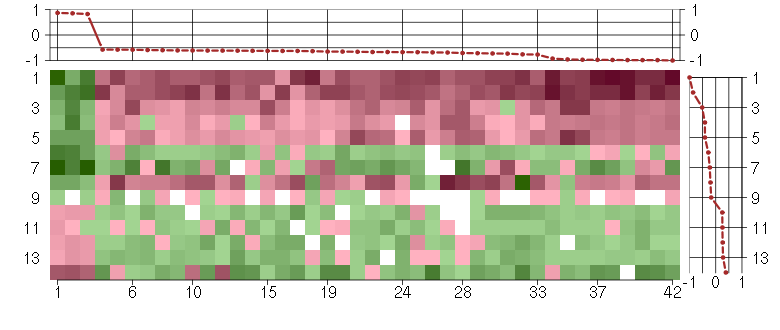

Under-expression is coded with green,
over-expression with red color.



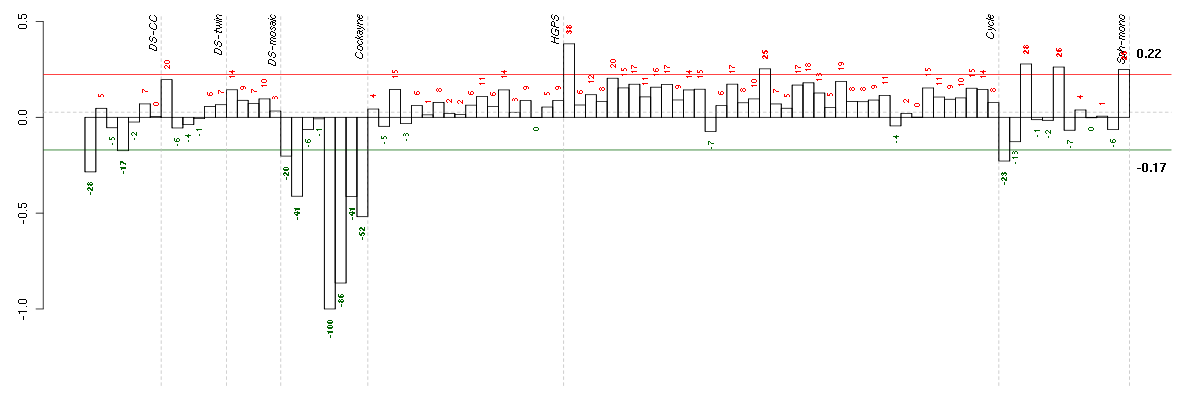
A2Malpha-2-macroglobulin (217757_at), score: -0.63 ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -0.96 AIM2absent in melanoma 2 (206513_at), score: -0.67 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: -0.57 CD24CD24 molecule (266_s_at), score: -0.67 CRTAPcartilage associated protein (201380_at), score: 0.87 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.71 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.62 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.67 FZD3frizzled homolog 3 (Drosophila) (219683_at), score: -0.61 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: -0.62 GAGE4G antigen 4 (207086_x_at), score: -0.98 GAGE6G antigen 6 (208155_x_at), score: -0.92 GATA3GATA binding protein 3 (209604_s_at), score: -0.99 HERC5hect domain and RLD 5 (219863_at), score: -0.77 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.82 HLA-DPA1major histocompatibility complex, class II, DP alpha 1 (211990_at), score: -0.62 HRASLSHRAS-like suppressor (219983_at), score: -0.61 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.67 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.69 NEFLneurofilament, light polypeptide (221805_at), score: -0.61 NMUneuromedin U (206023_at), score: -0.72 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.65 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.97 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.73 PDE10Aphosphodiesterase 10A (205501_at), score: -0.68 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.58 RAD23BRAD23 homolog B (S. cerevisiae) (201223_s_at), score: 0.85 REEP1receptor accessory protein 1 (204364_s_at), score: -0.6 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.66 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.71 TAC1tachykinin, precursor 1 (206552_s_at), score: -0.65 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.59 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.99 TMOD1tropomodulin 1 (203661_s_at), score: -0.61 TYRP1tyrosinase-related protein 1 (205694_at), score: -0.63 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.98 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -1 UGT1A8UDP glucuronosyltransferase 1 family, polypeptide A8 (221304_at), score: -0.63 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.97 WISP1WNT1 inducible signaling pathway protein 1 (206796_at), score: -0.57 WT1Wilms tumor 1 (206067_s_at), score: -0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |