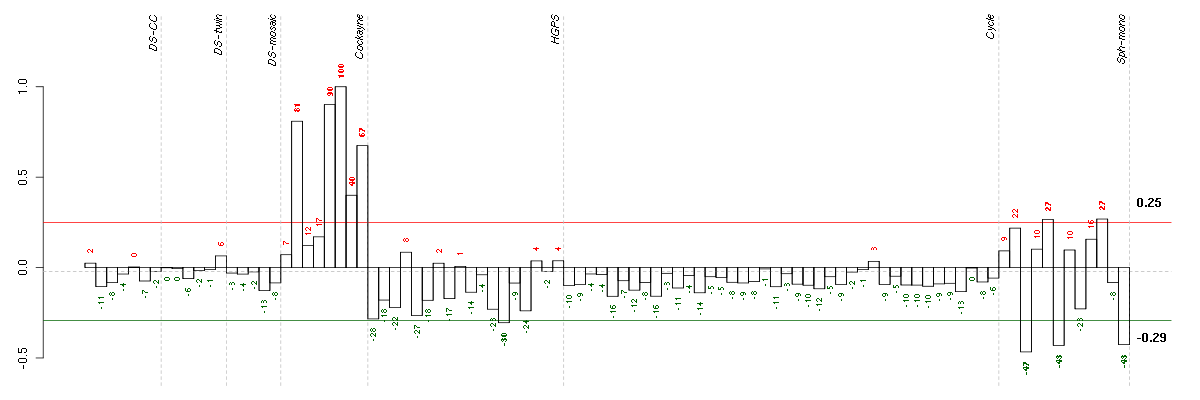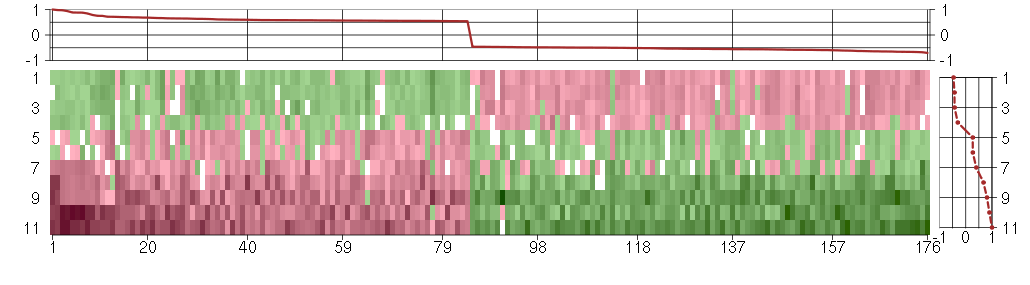

Under-expression is coded with green,
over-expression with red color.



ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: 1 ABHD14Aabhydrolase domain containing 14A (210006_at), score: -0.58 AGPSalkylglycerone phosphate synthase (205401_at), score: 0.55 AHNAK2AHNAK nucleoprotein 2 (212992_at), score: -0.49 AIM2absent in melanoma 2 (206513_at), score: 0.6 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: 0.58 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.63 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: 0.56 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (201043_s_at), score: 0.56 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: -0.46 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.56 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.57 ARSJarylsulfatase family, member J (219973_at), score: -0.48 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: 0.58 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: -0.63 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: -0.47 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.57 C4orf31chromosome 4 open reading frame 31 (219747_at), score: -0.56 C5orf30chromosome 5 open reading frame 30 (221823_at), score: -0.63 CCNG1cyclin G1 (208796_s_at), score: -0.51 CD24CD24 molecule (266_s_at), score: 0.72 CD248CD248 molecule, endosialin (219025_at), score: -0.56 CD81CD81 molecule (200675_at), score: -0.48 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: -0.57 COL11A1collagen, type XI, alpha 1 (37892_at), score: -0.64 CRATcarnitine acetyltransferase (209522_s_at), score: -0.48 CRTAPcartilage associated protein (201380_at), score: -0.58 CRYABcrystallin, alpha B (209283_at), score: -0.66 CTNNBL1catenin, beta like 1 (221021_s_at), score: 0.56 CTSKcathepsin K (202450_s_at), score: -0.49 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: 0.71 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.66 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (209178_at), score: 0.56 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.55 DIO2deiodinase, iodothyronine, type II (203700_s_at), score: 0.65 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.56 EPHB2EPH receptor B2 (209589_s_at), score: 0.69 EPS8L2EPS8-like 2 (218180_s_at), score: -0.49 FAM105Afamily with sequence similarity 105, member A (219694_at), score: 0.61 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: -0.47 FAM169Afamily with sequence similarity 169, member A (213954_at), score: 0.7 FARP1FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) (201911_s_at), score: -0.49 FASFas (TNF receptor superfamily, member 6) (204780_s_at), score: -0.59 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: -0.66 FBN2fibrillin 2 (203184_at), score: -0.5 FKBP4FK506 binding protein 4, 59kDa (200894_s_at), score: 0.68 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: -0.7 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: 0.55 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.61 GAGE4G antigen 4 (207086_x_at), score: 0.97 GAGE6G antigen 6 (208155_x_at), score: 0.94 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (219956_at), score: -0.49 GAS6growth arrest-specific 6 (202177_at), score: -0.58 GATA3GATA binding protein 3 (209604_s_at), score: 0.79 GHRgrowth hormone receptor (205498_at), score: -0.51 GLSglutaminase (203159_at), score: -0.62 GLTPglycolipid transfer protein (219267_at), score: -0.48 GPR124G protein-coupled receptor 124 (221814_at), score: -0.47 HAS2hyaluronan synthase 2 (206432_at), score: -0.56 HERC5hect domain and RLD 5 (219863_at), score: 0.75 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: -0.61 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: -0.46 HRASLSHRAS-like suppressor (219983_at), score: 0.57 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: -0.49 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: -0.63 HSPG2heparan sulfate proteoglycan 2 (201655_s_at), score: -0.49 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.56 IPO8importin 8 (205701_at), score: 0.58 IRS2insulin receptor substrate 2 (209185_s_at), score: -0.61 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.46 KIAA0895KIAA0895 (213424_at), score: 0.55 KRT19keratin 19 (201650_at), score: -0.5 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.55 LAMB3laminin, beta 3 (209270_at), score: 0.59 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: -0.52 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: -0.54 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.63 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: -0.52 LOC388796hypothetical LOC388796 (65588_at), score: 0.58 LOC728855hypothetical LOC728855 (222001_x_at), score: 0.69 LRP10low density lipoprotein receptor-related protein 10 (201412_at), score: -0.5 LRRC2leucine rich repeat containing 2 (219949_at), score: -0.65 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.6 LUMlumican (201744_s_at), score: -0.47 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: 0.55 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: -0.65 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: -0.53 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: 0.65 MFAP5microfibrillar associated protein 5 (213764_s_at), score: -0.65 MOXD1monooxygenase, DBH-like 1 (209708_at), score: -0.56 MRC2mannose receptor, C type 2 (37408_at), score: -0.5 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: 0.58 MXRA5matrix-remodelling associated 5 (209596_at), score: -0.48 MXRA8matrix-remodelling associated 8 (213422_s_at), score: -0.56 MYO1Cmyosin IC (214656_x_at), score: -0.5 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: 0.58 NDNnecdin homolog (mouse) (209550_at), score: -0.48 NEFLneurofilament, light polypeptide (221805_at), score: 0.67 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: -0.54 NFASCneurofascin homolog (chicken) (213438_at), score: -0.54 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: 0.57 NMUneuromedin U (206023_at), score: 0.63 NOC2Lnucleolar complex associated 2 homolog (S. cerevisiae) (202115_s_at), score: 0.54 NOL12nucleolar protein 12 (219324_at), score: 0.61 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: 0.65 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: 0.65 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: 0.98 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: -0.53 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: 0.71 PDE10Aphosphodiesterase 10A (205501_at), score: 0.57 PDGFCplatelet derived growth factor C (218718_at), score: -0.52 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: 0.67 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: 0.55 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: -0.55 PQBP1polyglutamine binding protein 1 (214527_s_at), score: 0.59 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: -0.46 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: -0.58 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.62 PSMC3proteasome (prosome, macropain) 26S subunit, ATPase, 3 (201267_s_at), score: 0.54 PSMD13proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 (201233_at), score: 0.57 PTGER3prostaglandin E receptor 3 (subtype EP3) (213933_at), score: -0.55 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: -0.54 RAB40BRAB40B, member RAS oncogene family (204547_at), score: -0.56 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: 0.57 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: -0.54 RAD23BRAD23 homolog B (S. cerevisiae) (201223_s_at), score: -0.68 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: 0.57 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: -0.5 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: -0.57 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: 0.64 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: 0.58 SAFB2scaffold attachment factor B2 (32099_at), score: 0.55 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: -0.54 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: 0.56 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: -0.58 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: -0.51 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: -0.55 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.6 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: 0.58 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: 0.6 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: 0.56 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: -0.49 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: 0.7 SSNA1Sjogren syndrome nuclear autoantigen 1 (210378_s_at), score: 0.54 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: -0.55 SSRP1structure specific recognition protein 1 (200956_s_at), score: 0.54 STSsteroid sulfatase (microsomal), isozyme S (203767_s_at), score: -0.53 SYTL2synaptotagmin-like 2 (220613_s_at), score: 0.68 TBC1D3TBC1 domain family, member 3 (209403_at), score: 0.58 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.59 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: 0.84 TEKTEK tyrosine kinase, endothelial (206702_at), score: -0.53 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: 0.56 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: -0.66 TMEM204transmembrane protein 204 (219315_s_at), score: -0.51 TMEM47transmembrane protein 47 (209656_s_at), score: -0.67 TMEM51transmembrane protein 51 (218815_s_at), score: 0.6 TMOD1tropomodulin 1 (203661_s_at), score: 0.65 TNS1tensin 1 (221748_s_at), score: -0.64 TOB1transducer of ERBB2, 1 (202704_at), score: -0.59 TRIM16tripartite motif-containing 16 (204341_at), score: 0.57 TSPAN13tetraspanin 13 (217979_at), score: -0.58 TSPYL1TSPY-like 1 (221493_at), score: -0.5 TSPYL5TSPY-like 5 (213122_at), score: -0.59 TUBG2tubulin, gamma 2 (203894_at), score: -0.48 TYRP1tyrosinase-related protein 1 (205694_at), score: 0.6 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.56 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: 0.56 UBL3ubiquitin-like 3 (201535_at), score: -0.66 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: 0.88 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: 0.88 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: 0.88 WDR74WD repeat domain 74 (221712_s_at), score: 0.58 WISP1WNT1 inducible signaling pathway protein 1 (206796_at), score: 0.55 WT1Wilms tumor 1 (206067_s_at), score: 0.75 ZNF281zinc finger protein 281 (218401_s_at), score: -0.47
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |