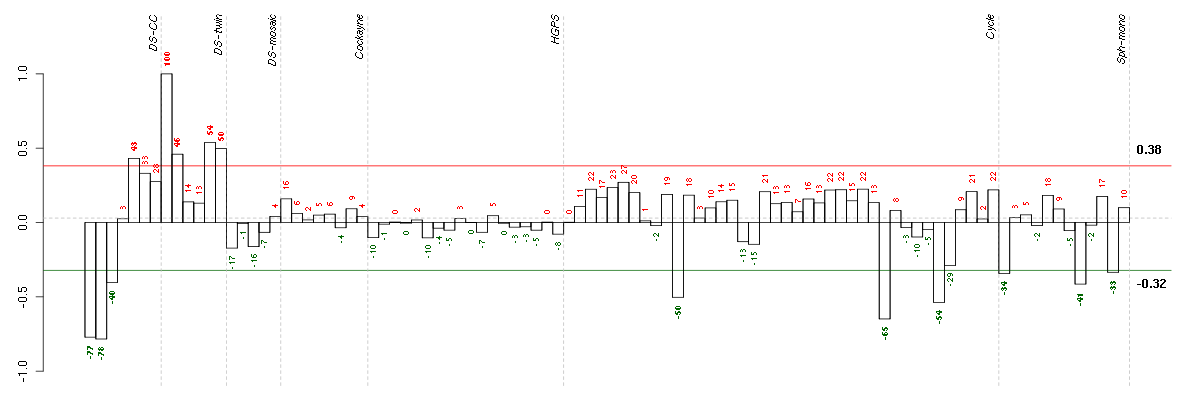

Under-expression is coded with green,
over-expression with red color.

response to molecule of bacterial origin
A change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cell proliferation
The multiplication or reproduction of cells, resulting in the expansion of a cell population.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
response to bacterium
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a bacterium.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
multi-organism process
Any process by which an organism has an effect on another organism of the same or different species.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
response to other organism
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from another living organism.
response to molecule of bacterial origin
A change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
response to molecule of bacterial origin
A change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
cytokine activity
Functions to control the survival, growth, differentiation and effector function of tissues and cells.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 9.059e-03 | 0.5368 | 6 | 67 | Cytokine-cytokine receptor interaction |
ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: -0.69 ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: -0.63 ANGPTL4angiopoietin-like 4 (221009_s_at), score: -0.81 APODapolipoprotein D (201525_at), score: -0.79 BMP2bone morphogenetic protein 2 (205289_at), score: -0.72 CD302CD302 molecule (203799_at), score: -0.59 COL15A1collagen, type XV, alpha 1 (203477_at), score: -0.66 CXCL2chemokine (C-X-C motif) ligand 2 (209774_x_at), score: -0.71 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: -0.9 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: -0.92 DPTdermatopontin (213068_at), score: -0.62 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: -0.81 EGR3early growth response 3 (206115_at), score: -0.75 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: -0.97 GCH1GTP cyclohydrolase 1 (204224_s_at), score: -0.61 GEMGTP binding protein overexpressed in skeletal muscle (204472_at), score: -0.65 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: -0.62 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: -0.69 HK2hexokinase 2 (202934_at), score: -0.58 IL11interleukin 11 (206924_at), score: -0.58 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: -0.57 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: -0.79 IL6interleukin 6 (interferon, beta 2) (205207_at), score: -0.72 IL8interleukin 8 (211506_s_at), score: -0.76 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: -0.58 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: -0.86 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: -1 KIAA1024KIAA1024 (215081_at), score: -0.66 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: -0.68 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: -0.62 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: -0.73 MFAP4microfibrillar-associated protein 4 (212713_at), score: -0.6 NAMPTnicotinamide phosphoribosyltransferase (217738_at), score: -0.61 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: -0.73 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: -0.93 PDGFDplatelet derived growth factor D (219304_s_at), score: -0.6 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: -0.75 PPLperiplakin (203407_at), score: -0.78 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: -0.6 RRADRas-related associated with diabetes (204802_at), score: -0.83 RUNX1runt-related transcription factor 1 (209360_s_at), score: -0.57 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (209681_at), score: -0.7 SMOXspermine oxidase (210357_s_at), score: -0.78 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: -0.63 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: -0.71 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: -0.63 STK38Lserine/threonine kinase 38 like (212572_at), score: -0.62 TBX3T-box 3 (219682_s_at), score: -0.64 THBDthrombomodulin (203887_s_at), score: -0.84 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: -0.59 VEGFAvascular endothelial growth factor A (211527_x_at), score: -0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |