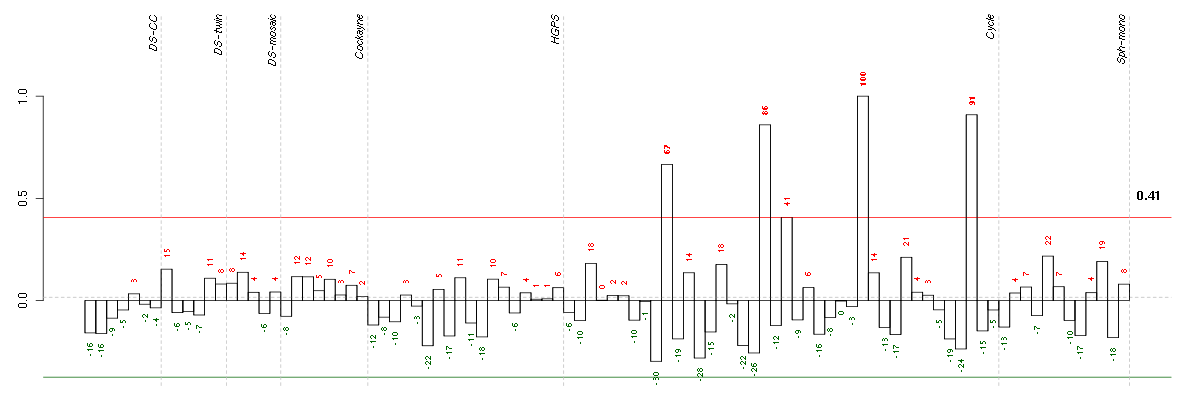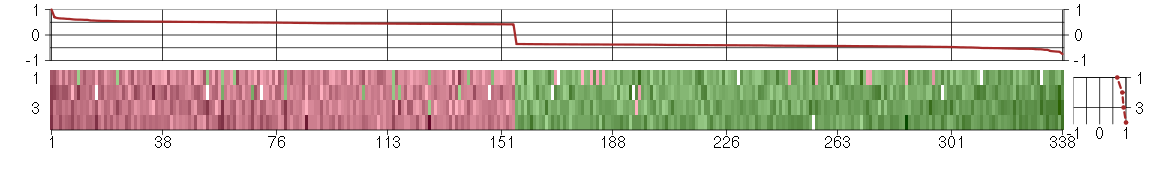

Under-expression is coded with green,
over-expression with red color.



ABCA2ATP-binding cassette, sub-family A (ABC1), member 2 (212772_s_at), score: -0.37 ACANaggrecan (205679_x_at), score: 0.52 ADAM21ADAM metallopeptidase domain 21 (207665_at), score: 0.51 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.47 ADAMTSL4ADAMTS-like 4 (220578_at), score: -0.36 ADSSadenylosuccinate synthase (221761_at), score: -0.37 AFF2AF4/FMR2 family, member 2 (206105_at), score: -0.44 AFG3L2AFG3 ATPase family gene 3-like 2 (yeast) (202486_at), score: 0.44 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.54 AHI1Abelson helper integration site 1 (221569_at), score: -0.43 ANXA10annexin A10 (210143_at), score: -0.56 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: -0.45 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.52 ARL17ADP-ribosylation factor-like 17 (210435_at), score: -0.45 ARMC9armadillo repeat containing 9 (219637_at), score: 0.51 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.53 ARSBarylsulfatase B (206129_s_at), score: 0.44 ASPNasporin (219087_at), score: -0.44 ASTN2astrotactin 2 (209693_at), score: 0.44 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: 0.46 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.6 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: -0.41 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: -0.36 BAXBCL2-associated X protein (208478_s_at), score: 0.52 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: 0.44 BCL2B-cell CLL/lymphoma 2 (203685_at), score: -0.45 BTBD3BTB (POZ) domain containing 3 (202946_s_at), score: -0.38 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: 0.46 C11orf51chromosome 11 open reading frame 51 (204218_at), score: 0.42 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: 0.49 C12orf52chromosome 12 open reading frame 52 (221777_at), score: 0.51 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.6 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.53 C21orf91chromosome 21 open reading frame 91 (220941_s_at), score: 0.45 C2CD2LC2CD2-like (204757_s_at), score: -0.45 C2orf49chromosome 2 open reading frame 49 (219662_at), score: 0.42 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.51 C8orf41chromosome 8 open reading frame 41 (219124_at), score: 0.5 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.56 CA12carbonic anhydrase XII (203963_at), score: 0.45 CALB1calbindin 1, 28kDa (205626_s_at), score: -0.39 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.43 CAPN3calpain 3, (p94) (210944_s_at), score: -0.38 CBLBCas-Br-M (murine) ecotropic retroviral transforming sequence b (209682_at), score: -0.4 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.47 CCNT2cyclin T2 (204645_at), score: -0.44 CD320CD320 molecule (218529_at), score: 0.65 CD40CD40 molecule, TNF receptor superfamily member 5 (35150_at), score: -0.36 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.52 CD83CD83 molecule (204440_at), score: -0.43 CDCA4cell division cycle associated 4 (218399_s_at), score: -0.37 CDCP1CUB domain containing protein 1 (218451_at), score: -0.43 CDH18cadherin 18, type 2 (206280_at), score: 0.43 CDK5cyclin-dependent kinase 5 (204247_s_at), score: 0.47 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: 0.43 CEP70centrosomal protein 70kDa (219036_at), score: 0.42 CHD2chromodomain helicase DNA binding protein 2 (203461_at), score: -0.38 CHRM2cholinergic receptor, muscarinic 2 (221330_at), score: 0.45 CLCA2chloride channel accessory 2 (206165_s_at), score: -0.46 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.44 CLEC1AC-type lectin domain family 1, member A (219761_at), score: -0.4 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.64 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: -0.38 CLK1CDC-like kinase 1 (214683_s_at), score: -0.47 CLK4CDC-like kinase 4 (210346_s_at), score: -0.39 CLUclusterin (208791_at), score: -0.51 CNNM2cyclin M2 (206818_s_at), score: 0.52 CNPY3canopy 3 homolog (zebrafish) (217931_at), score: 0.44 COASYCoenzyme A synthase (201913_s_at), score: 0.43 COG2component of oligomeric golgi complex 2 (203073_at), score: 0.45 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.46 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.65 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.43 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: 0.49 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.51 CPT1Acarnitine palmitoyltransferase 1A (liver) (203633_at), score: 0.53 CRYL1crystallin, lambda 1 (220753_s_at), score: -0.47 CST6cystatin E/M (206595_at), score: -0.54 CSTAcystatin A (stefin A) (204971_at), score: -0.36 CTDP1CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 (205035_at), score: 0.42 CTSEcathepsin E (205927_s_at), score: -0.4 CTSKcathepsin K (202450_s_at), score: -0.45 CTSOcathepsin O (203758_at), score: -0.44 CTSScathepsin S (202901_x_at), score: -0.37 CTSZcathepsin Z (210042_s_at), score: 0.42 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.44 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (216809_at), score: -0.38 CYP11B1cytochrome P450, family 11, subfamily B, polypeptide 1 (214610_at), score: -0.36 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: -0.39 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: 0.47 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.43 DCPSdecapping enzyme, scavenger (218774_at), score: 0.44 DHRS2dehydrogenase/reductase (SDR family) member 2 (214079_at), score: -0.39 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.58 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.37 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: -0.45 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: 0.57 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.45 DUSP6dual specificity phosphatase 6 (208891_at), score: -0.51 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.45 EDNRBendothelin receptor type B (204271_s_at), score: -0.49 EGFL6EGF-like-domain, multiple 6 (219454_at), score: -0.42 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: -0.4 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: -0.39 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: -0.41 EMCNendomucin (219436_s_at), score: 0.63 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.58 EML4echinoderm microtubule associated protein like 4 (220386_s_at), score: 0.47 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.4 EPOerythropoietin (217254_s_at), score: 0.44 EREGepiregulin (205767_at), score: -0.43 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.39 ESRRAestrogen-related receptor alpha (1487_at), score: -0.4 EVI2Aecotropic viral integration site 2A (204774_at), score: 0.52 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.52 EXOSC5exosome component 5 (218481_at), score: 0.51 FAM13Bfamily with sequence similarity 13, member B (218518_at), score: -0.38 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.39 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: -0.39 FAM48Afamily with sequence similarity 48, member A (221774_x_at), score: 0.44 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.42 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.61 FBXO31F-box protein 31 (219785_s_at), score: 0.52 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: -0.36 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: -0.43 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.55 GAAglucosidase, alpha; acid (202812_at), score: -0.48 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: 0.54 GANgigaxonin (220124_at), score: -0.44 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: -0.37 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.42 GKglycerol kinase (207387_s_at), score: -0.46 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: 0.54 GNPDA1glucosamine-6-phosphate deaminase 1 (202382_s_at), score: 0.42 GPM6Bglycoprotein M6B (209167_at), score: -0.5 GPR153G protein-coupled receptor 153 (221902_at), score: -0.44 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.51 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.65 GPSM3G-protein signaling modulator 3 (AGS3-like, C. elegans) (204265_s_at), score: 0.49 GPSN2glycoprotein, synaptic 2 (208336_s_at), score: 0.45 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.36 GRLF1glucocorticoid receptor DNA binding factor 1 (202044_at), score: 0.43 HBBhemoglobin, beta (211696_x_at), score: -0.46 HECTD3HECT domain containing 3 (218632_at), score: 0.43 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.53 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: 0.51 HOMER2homer homolog 2 (Drosophila) (217080_s_at), score: -0.37 HOXD1homeobox D1 (205975_s_at), score: -0.41 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.6 HTN1histatin 1 (206639_x_at), score: -0.41 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.43 IER2immediate early response 2 (202081_at), score: -0.41 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: -0.53 IL12Ainterleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) (207160_at), score: -0.38 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.53 IL33interleukin 33 (209821_at), score: -0.75 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.4 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.55 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: -0.37 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: -0.4 KIAA0415KIAA0415 (209912_s_at), score: 0.53 KIAA0586KIAA0586 (205631_at), score: 0.52 KIAA1024KIAA1024 (215081_at), score: -0.41 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.51 KLHL2kelch-like 2, Mayven (Drosophila) (219157_at), score: -0.37 KRT33Akeratin 33A (208483_x_at), score: -0.51 KRT9keratin 9 (208188_at), score: -0.43 LANCL2LanC lantibiotic synthetase component C-like 2 (bacterial) (218219_s_at), score: 0.46 LHX3LIM homeobox 3 (221670_s_at), score: -0.37 LIG3ligase III, DNA, ATP-dependent (204123_at), score: 0.46 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: 0.5 LOC100128223hypothetical protein LOC100128223 (221264_s_at), score: 0.43 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.49 LOC92249hypothetical LOC92249 (212957_s_at), score: 0.44 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200785_s_at), score: -0.37 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.43 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.36 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: -0.35 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.35 MAP4K4mitogen-activated protein kinase kinase kinase kinase 4 (218181_s_at), score: -0.39 MAP7D3MAP7 domain containing 3 (219576_at), score: 0.44 MASP2mannan-binding lectin serine peptidase 2 (207041_at), score: -0.42 MBmyoglobin (204179_at), score: -0.37 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: -0.38 MLPHmelanophilin (218211_s_at), score: 0.59 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.38 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.46 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.46 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (220346_at), score: -0.37 MTHFSDmethenyltetrahydrofolate synthetase domain containing (218879_s_at), score: 0.47 MTMR1myotubularin related protein 1 (213511_s_at), score: -0.4 MTMR9myotubularin related protein 9 (204837_at), score: -0.41 MUSKmuscle, skeletal, receptor tyrosine kinase (207633_s_at), score: -0.44 MXRA5matrix-remodelling associated 5 (209596_at), score: -0.37 NAGAN-acetylgalactosaminidase, alpha- (202943_s_at), score: 0.44 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: 0.53 NARS2asparaginyl-tRNA synthetase 2, mitochondrial (putative) (219217_at), score: 0.45 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: -0.4 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.49 NEFLneurofilament, light polypeptide (221805_at), score: -0.63 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.54 NMBneuromedin B (205204_at), score: 0.53 NOL8nucleolar protein 8 (218244_at), score: 0.51 NR0B1nuclear receptor subfamily 0, group B, member 1 (206644_at), score: -0.35 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.41 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.56 NTNG1netrin G1 (206713_at), score: -0.49 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.45 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: 0.5 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (205552_s_at), score: -0.38 OLFM1olfactomedin 1 (213131_at), score: -0.37 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.52 OMDosteomodulin (205907_s_at), score: -0.38 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.43 OR2W1olfactory receptor, family 2, subfamily W, member 1 (221451_s_at), score: -0.38 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.53 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: 0.43 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.47 PENKproenkephalin (213791_at), score: -0.43 PET112LPET112-like (yeast) (204300_at), score: 0.43 PFDN6prefoldin subunit 6 (222029_x_at), score: 0.45 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.43 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.53 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.48 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.45 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.38 PLEKHA2pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 (217677_at), score: 0.48 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204285_s_at), score: -0.54 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.54 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: 0.51 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.44 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: -0.43 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.66 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.4 PRSS3protease, serine, 3 (213421_x_at), score: -0.56 PSD4pleckstrin and Sec7 domain containing 4 (203317_at), score: -0.45 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: 0.46 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.49 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: 0.5 PTENP1phosphatase and tensin homolog pseudogene 1 (217492_s_at), score: 0.42 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.54 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.4 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.42 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: 0.59 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (219112_at), score: -0.41 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: -0.47 RASA2RAS p21 protein activator 2 (206636_at), score: -0.42 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: -0.47 RBM39RNA binding motif protein 39 (207941_s_at), score: -0.49 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.46 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.44 RFX7regulatory factor X, 7 (218430_s_at), score: 0.51 RNF2ring finger protein 2 (205215_at), score: -0.41 RNF44ring finger protein 44 (203286_at), score: 0.49 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.48 RPL14ribosomal protein L14 (219138_at), score: 0.45 RPL23ribosomal protein L23 (214744_s_at), score: -0.42 RPL27Aribosomal protein L27a (212044_s_at), score: 0.48 RPS11ribosomal protein S11 (213350_at), score: 0.5 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.54 SCAPERS phase cyclin A-associated protein in the ER (209741_x_at), score: -0.38 SEC14L5SEC14-like 5 (S. cerevisiae) (210169_at), score: -0.43 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.45 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.49 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.63 SEZ6L2seizure related 6 homolog (mouse)-like 2 (218720_x_at), score: -0.36 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: -0.46 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.55 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: -0.42 SLC17A6solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (220551_at), score: -0.4 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: -0.44 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: -0.39 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (211754_s_at), score: -0.42 SLC26A4solute carrier family 26, member 4 (206529_x_at), score: -0.37 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.69 SLKSTE20-like kinase (yeast) (206875_s_at), score: -0.42 SMU1smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) (218393_s_at), score: 0.46 SNX13sorting nexin 13 (213292_s_at), score: 0.54 SORT1sortilin 1 (212807_s_at), score: 0.46 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.45 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.52 SPP1secreted phosphoprotein 1 (209875_s_at), score: 0.52 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.37 SSFA2sperm specific antigen 2 (202506_at), score: -0.38 STC1stanniocalcin 1 (204597_x_at), score: -0.4 STK16serine/threonine kinase 16 (209622_at), score: 0.51 STOMstomatin (201060_x_at), score: 0.42 STRA6stimulated by retinoic acid gene 6 homolog (mouse) (221701_s_at), score: -0.36 SUOXsulfite oxidase (204067_at), score: 0.45 SYNMsynemin, intermediate filament protein (212730_at), score: 0.54 SYT1synaptotagmin I (203999_at), score: -0.4 SYT11synaptotagmin XI (209198_s_at), score: 0.57 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: 0.44 TBX5T-box 5 (207155_at), score: 0.48 TCF7L1transcription factor 7-like 1 (T-cell specific, HMG-box) (221016_s_at), score: 0.46 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.52 TGFAtransforming growth factor, alpha (205016_at), score: -0.43 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (205210_at), score: 0.5 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: 0.44 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.48 TIMM9translocase of inner mitochondrial membrane 9 homolog (yeast) (218316_at), score: 0.49 TMEM158transmembrane protein 158 (213338_at), score: -0.42 TMEM160transmembrane protein 160 (219219_at), score: 0.49 TMOD1tropomodulin 1 (203661_s_at), score: -0.38 TMSB15Athymosin beta 15a (205347_s_at), score: 0.66 TNIKTRAF2 and NCK interacting kinase (213107_at), score: 0.45 TOMM22translocase of outer mitochondrial membrane 22 homolog (yeast) (217960_s_at), score: 0.49 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.47 TP53TG1TP53 target 1 (non-protein coding) (209917_s_at), score: 0.49 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.56 TRIAP1TP53 regulated inhibitor of apoptosis 1 (218403_at), score: 0.48 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: 0.49 TRPC1transient receptor potential cation channel, subfamily C, member 1 (205802_at), score: -0.46 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.37 TRY6trypsinogen C (215395_x_at), score: -0.42 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (214755_at), score: 0.44 UCHL5IPUCHL5 interacting protein (213334_x_at), score: 0.44 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.42 USE1unconventional SNARE in the ER 1 homolog (S. cerevisiae) (221706_s_at), score: 0.45 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.48 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.36 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.5 WDR52WD repeat domain 52 (221103_s_at), score: -0.37 XPO4exportin 4 (218479_s_at), score: 0.42 YIPF1Yip1 domain family, member 1 (214733_s_at), score: 0.45 YY2YY2 transcription factor (216531_at), score: 0.51 ZNF174zinc finger protein 174 (210291_s_at), score: 0.46 ZNF195zinc finger protein 195 (204234_s_at), score: 0.43 ZNF219zinc finger protein 219 (219314_s_at), score: -0.5 ZNF22zinc finger protein 22 (KOX 15) (218005_at), score: 0.46 ZNF236zinc finger protein 236 (47571_at), score: -0.39 ZNF350zinc finger protein 350 (219266_at), score: 0.43 ZNF434zinc finger protein 434 (218937_at), score: 0.42 ZNF667zinc finger protein 667 (207120_at), score: 0.53 ZNF688zinc finger protein 688 (213527_s_at), score: 0.45 ZNF749zinc finger protein 749 (215289_at), score: -0.52
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |