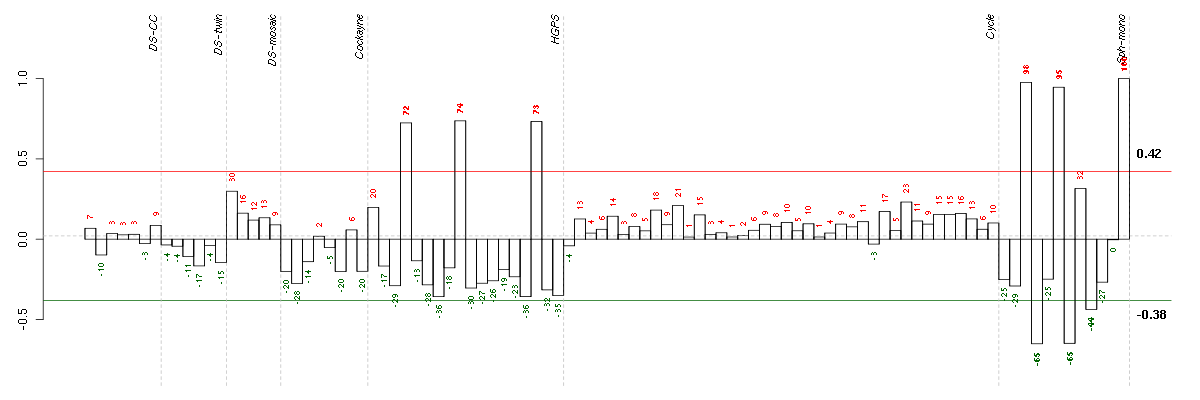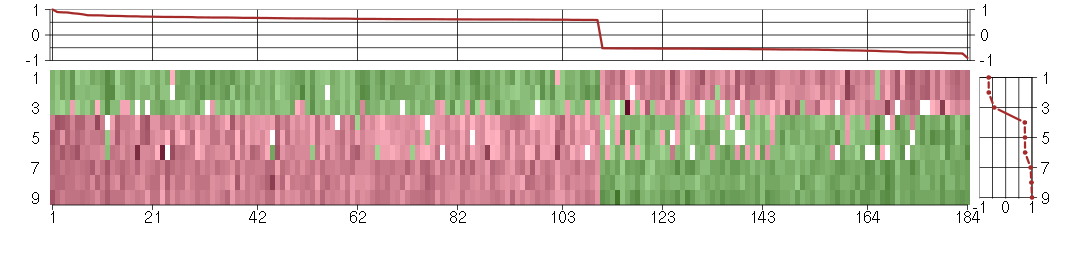

Under-expression is coded with green,
over-expression with red color.



ABHD2abhydrolase domain containing 2 (221815_at), score: -0.56 ACTG2actin, gamma 2, smooth muscle, enteric (202274_at), score: 0.64 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: 0.61 AEBP1AE binding protein 1 (201792_at), score: 0.64 AGTR1angiotensin II receptor, type 1 (205357_s_at), score: 0.6 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: -0.53 AOX1aldehyde oxidase 1 (205083_at), score: 0.61 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.62 ARID5BAT rich interactive domain 5B (MRF1-like) (212614_at), score: 0.61 ARL15ADP-ribosylation factor-like 15 (219842_at), score: 0.68 ARSJarylsulfatase family, member J (219973_at), score: 0.62 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (212135_s_at), score: 0.69 AXLAXL receptor tyrosine kinase (202685_s_at), score: 0.62 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: -0.53 BGNbiglycan (213905_x_at), score: 0.65 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.65 C19orf2chromosome 19 open reading frame 2 (214173_x_at), score: -0.57 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: 0.7 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.61 C9orf95chromosome 9 open reading frame 95 (219147_s_at), score: 0.65 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: -0.68 CAPNS1calpain, small subunit 1 (200001_at), score: 0.6 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: -0.56 CCDC92coiled-coil domain containing 92 (218175_at), score: 0.7 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.69 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.71 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: -0.53 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.62 CIRCBF1 interacting corepressor (209571_at), score: -0.53 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.9 CNTNAP1contactin associated protein 1 (219400_at), score: 0.68 COBLL1COBL-like 1 (203642_s_at), score: 0.78 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: 0.64 CORINcorin, serine peptidase (220356_at), score: 0.71 CREB3L1cAMP responsive element binding protein 3-like 1 (213059_at), score: 0.6 CROTcarnitine O-octanoyltransferase (204573_at), score: -0.53 CRYABcrystallin, alpha B (209283_at), score: 0.62 CSRP2cysteine and glycine-rich protein 2 (211126_s_at), score: 0.63 CTGFconnective tissue growth factor (209101_at), score: 0.61 CYB561cytochrome b-561 (209163_at), score: -0.57 CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: 0.6 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.72 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.65 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.55 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.55 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.62 DIDO1death inducer-obliterator 1 (218325_s_at), score: -0.56 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.84 EHD3EH-domain containing 3 (218935_at), score: 0.67 EIF1eukaryotic translation initiation factor 1 (212130_x_at), score: -0.54 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: -0.72 EPHB6EPH receptor B6 (204718_at), score: 0.61 EREGepiregulin (205767_at), score: 0.65 EXT1exostoses (multiple) 1 (201995_at), score: 0.75 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: -0.58 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.66 FIG4FIG4 homolog (S. cerevisiae) (203656_at), score: 0.62 FLNAfilamin A, alpha (actin binding protein 280) (213746_s_at), score: 0.59 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: 0.62 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.75 FOXL2forkhead box L2 (220102_at), score: -0.55 FRYfurry homolog (Drosophila) (204072_s_at), score: 0.62 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.62 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.9 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.62 GLRBglycine receptor, beta (205280_at), score: 0.59 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.72 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.75 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.67 GTDC1glycosyltransferase-like domain containing 1 (219770_at), score: 0.73 HERC2P2hect domain and RLD 2 pseudogene 2 (217317_s_at), score: -0.53 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.65 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: -0.53 HTATIP2HIV-1 Tat interactive protein 2, 30kDa (209448_at), score: 0.64 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.71 ILKintegrin-linked kinase (201234_at), score: 0.59 IMAASLC7A5 pseudogene (208118_x_at), score: -0.69 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.77 IQCKIQ motif containing K (213392_at), score: -0.7 ISL1ISL LIM homeobox 1 (206104_at), score: 0.62 KANK2KN motif and ankyrin repeat domains 2 (218418_s_at), score: 0.61 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: -0.54 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: -0.54 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.64 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.59 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.7 LIN7Alin-7 homolog A (C. elegans) (206440_at), score: 0.61 LMO7LIM domain 7 (202674_s_at), score: 0.61 LOC440354PI-3-kinase-related kinase SMG-1 pseudogene (210396_s_at), score: -0.52 LPPR4plasticity related gene 1 (213496_at), score: 0.88 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: -0.61 LRRC16Aleucine rich repeat containing 16A (219573_at), score: 0.71 LUMlumican (201744_s_at), score: 0.71 MAB21L1mab-21-like 1 (C. elegans) (206163_at), score: 0.65 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: -0.68 MAMLD1mastermind-like domain containing 1 (205088_at), score: 0.63 MANSC1MANSC domain containing 1 (220945_x_at), score: 0.74 MAP1Amicrotubule-associated protein 1A (203151_at), score: 0.63 ME1malic enzyme 1, NADP(+)-dependent, cytosolic (204059_s_at), score: 0.61 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: 0.6 MEFVMediterranean fever (208262_x_at), score: -0.6 MICAL2microtubule associated monoxygenase, calponin and LIM domain containing 2 (212473_s_at), score: 0.63 MLF1myeloid leukemia factor 1 (204784_s_at), score: -0.57 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: 0.6 MMDmonocyte to macrophage differentiation-associated (203414_at), score: -0.67 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.64 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: -0.6 MYL9myosin, light chain 9, regulatory (201058_s_at), score: 0.59 MYO1Bmyosin IB (212364_at), score: 0.69 MYO1Emyosin IE (203072_at), score: 0.65 NAV3neuron navigator 3 (204823_at), score: 0.77 NBEAneurobeachin (221207_s_at), score: 0.85 NBPF10neuroblastoma breakpoint family, member 10 (214693_x_at), score: -0.54 NEDD9neural precursor cell expressed, developmentally down-regulated 9 (202149_at), score: 0.6 NESnestin (218678_at), score: 0.81 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: 0.61 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: 0.61 OPTNoptineurin (202073_at), score: 0.61 OXTRoxytocin receptor (206825_at), score: 0.64 PCGF1polycomb group ring finger 1 (210023_s_at), score: -0.52 PDE8Aphosphodiesterase 8A (212522_at), score: -0.53 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.69 PDLIM1PDZ and LIM domain 1 (208690_s_at), score: 0.72 PER2period homolog 2 (Drosophila) (205251_at), score: -0.54 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.64 PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: -0.53 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.66 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.63 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.55 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: -0.68 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: 0.68 PRSS3protease, serine, 3 (213421_x_at), score: 0.66 PTPN13protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) (204201_s_at), score: 0.62 RGNEFRho-guanine nucleotide exchange factor (219610_at), score: -0.65 RPL5ribosomal protein L5 (213689_x_at), score: 0.63 S100A4S100 calcium binding protein A4 (203186_s_at), score: 0.67 SAP30BPSAP30 binding protein (217965_s_at), score: -0.54 SAPS2SAPS domain family, member 2 (202792_s_at), score: -0.55 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.57 SCD5stearoyl-CoA desaturase 5 (220232_at), score: -0.55 SCN3Asodium channel, voltage-gated, type III, alpha subunit (210432_s_at), score: 0.77 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: 0.64 SEPP1selenoprotein P, plasma, 1 (201427_s_at), score: -0.57 SH3BGRL3SH3 domain binding glutamic acid-rich protein like 3 (221269_s_at), score: 0.63 SKP1S-phase kinase-associated protein 1 (200719_at), score: -0.56 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: -0.6 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: -0.56 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.53 SNNstannin (218032_at), score: -0.72 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.57 SPATA2spermatogenesis associated 2 (204433_s_at), score: -0.53 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: -0.53 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.68 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.73 SYNGR1synaptogyrin 1 (210613_s_at), score: 0.63 SYNMsynemin, intermediate filament protein (212730_at), score: 0.89 SYT11synaptotagmin XI (209198_s_at), score: 0.71 TAGLNtransgelin (205547_s_at), score: 0.61 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.6 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.58 TBXA2Rthromboxane A2 receptor (336_at), score: 1 TCF7transcription factor 7 (T-cell specific, HMG-box) (205255_x_at), score: -0.53 TEAD3TEA domain family member 3 (209454_s_at), score: 0.72 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: -0.58 TGFB1I1transforming growth factor beta 1 induced transcript 1 (209651_at), score: 0.68 THSD1thrombospondin, type I, domain containing 1 (219477_s_at), score: -0.68 TIMM22translocase of inner mitochondrial membrane 22 homolog (yeast) (219184_x_at), score: 0.61 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: -0.53 TMEM111transmembrane protein 111 (217882_at), score: -0.59 TMEM2transmembrane protein 2 (218113_at), score: 0.62 TMEM209transmembrane protein 209 (222267_at), score: -0.56 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.73 TNS1tensin 1 (221748_s_at), score: 0.67 TRAF5TNF receptor-associated factor 5 (204352_at), score: 0.67 TRY6trypsinogen C (215395_x_at), score: 0.71 UGDHUDP-glucose dehydrogenase (203343_at), score: 0.69 UGP2UDP-glucose pyrophosphorylase 2 (205480_s_at), score: 0.59 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.67 WASLWiskott-Aldrich syndrome-like (205809_s_at), score: -0.53 WWP2WW domain containing E3 ubiquitin protein ligase 2 (204022_at), score: 0.61 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: -0.61 ZNF212zinc finger protein 212 (203985_at), score: -0.52 ZNF273zinc finger protein 273 (215239_x_at), score: -0.57 ZYXzyxin (200808_s_at), score: 0.61
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |