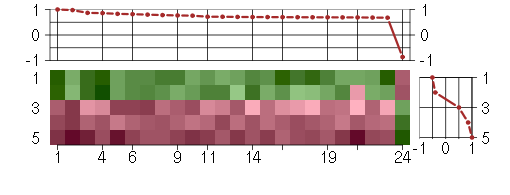

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: 0.7 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 1 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 0.98 CH25Hcholesterol 25-hydroxylase (206932_at), score: 0.69 CKBcreatine kinase, brain (200884_at), score: 0.77 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.78 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.82 EMX2empty spiracles homeobox 2 (221950_at), score: 0.69 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.72 IER3immediate early response 3 (201631_s_at), score: -0.86 INHBBinhibin, beta B (205258_at), score: 0.71 IRX5iroquois homeobox 5 (210239_at), score: 0.87 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: 0.72 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.7 NCAM1neural cell adhesion molecule 1 (212843_at), score: 0.69 NTF3neurotrophin 3 (206706_at), score: 0.68 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.7 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: 0.8 RASIP1Ras interacting protein 1 (220027_s_at), score: 0.76 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.69 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.83 TBX5T-box 5 (207155_at), score: 0.68 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.86 TSPAN13tetraspanin 13 (217979_at), score: 0.7
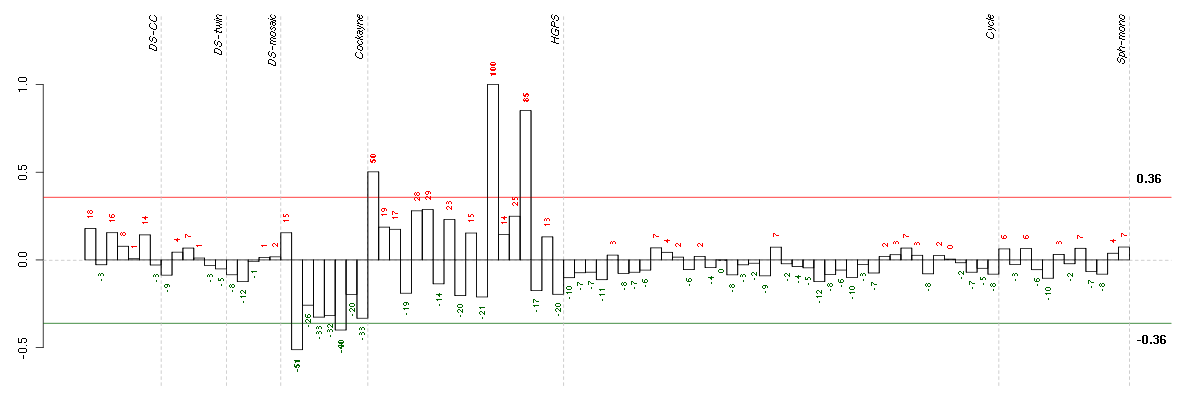
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |