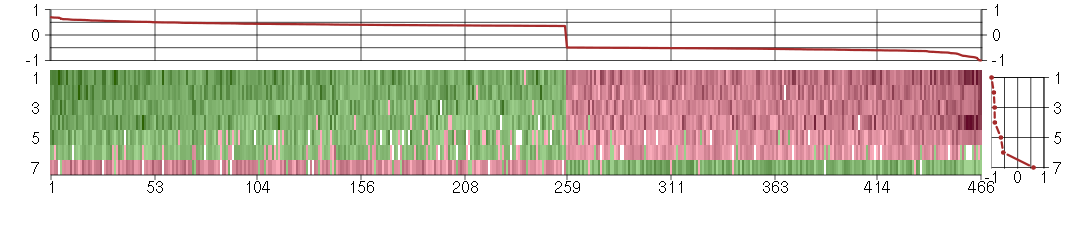

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
ectoderm development
The process whose specific outcome is the progression of the ectoderm over time, from its formation to the mature structure. In animal embryos, the ectoderm is the outer germ layer of the embryo, formed during gastrulation.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
epidermis development
The process whose specific outcome is the progression of the epidermis over time, from its formation to the mature structure. The epidermis is the outer epithelial layer of a plant or animal, it may be a single layer that produces an extracellular material (e.g. the cuticle of arthropods) or a complex stratified squamous epithelium, as in the case of many vertebrate species.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
epidermis development
The process whose specific outcome is the progression of the epidermis over time, from its formation to the mature structure. The epidermis is the outer epithelial layer of a plant or animal, it may be a single layer that produces an extracellular material (e.g. the cuticle of arthropods) or a complex stratified squamous epithelium, as in the case of many vertebrate species.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

A2Malpha-2-macroglobulin (217757_at), score: -0.57 ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -1 ABHD14Aabhydrolase domain containing 14A (210006_at), score: 0.44 ACIN1apoptotic chromatin condensation inducer 1 (201715_s_at), score: -0.67 ACP2acid phosphatase 2, lysosomal (202767_at), score: 0.36 ACSL3acyl-CoA synthetase long-chain family member 3 (201660_at), score: 0.47 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: 0.45 ACYP2acylphosphatase 2, muscle type (206833_s_at), score: 0.37 ADORA1adenosine A1 receptor (216220_s_at), score: 0.39 AGPSalkylglycerone phosphate synthase (205401_at), score: -0.54 AHNAK2AHNAK nucleoprotein 2 (212992_at), score: 0.4 AIM2absent in melanoma 2 (206513_at), score: -0.68 AIParyl hydrocarbon receptor interacting protein (201782_s_at), score: -0.52 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: -0.63 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: 0.41 ANGPTL2angiopoietin-like 2 (213004_at), score: -0.53 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (201043_s_at), score: -0.61 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: -0.54 APCadenomatous polyposis coli (203525_s_at), score: 0.4 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.36 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: -0.54 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: -0.55 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.61 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (216266_s_at), score: -0.5 ARHGAP1Rho GTPase activating protein 1 (202117_at), score: 0.42 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.44 ARPP-19cyclic AMP phosphoprotein, 19 kD (214553_s_at), score: -0.51 ARSAarylsulfatase A (204443_at), score: 0.36 ASXL2additional sex combs like 2 (Drosophila) (218659_at), score: -0.52 ATP11BATPase, class VI, type 11B (212536_at), score: 0.36 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: -0.6 ATP6V1FATPase, H+ transporting, lysosomal 14kDa, V1 subunit F (201527_at), score: -0.52 ATP9AATPase, class II, type 9A (212062_at), score: 0.38 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: 0.37 B4GALT7xylosylprotein beta 1,4-galactosyltransferase, polypeptide 7 (galactosyltransferase I) (53076_at), score: 0.35 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: 0.53 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: -0.52 BAT1HLA-B associated transcript 1 (200041_s_at), score: -0.5 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: -0.72 BCKDKbranched chain ketoacid dehydrogenase kinase (202030_at), score: 0.45 BCLAF1BCL2-associated transcription factor 1 (201101_s_at), score: -0.5 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: -0.69 BICD2bicaudal D homolog 2 (Drosophila) (209203_s_at), score: -0.51 BTRCbeta-transducin repeat containing (216091_s_at), score: -0.5 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.6 C11orf17chromosome 11 open reading frame 17 (219953_s_at), score: -0.56 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: -0.5 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.4 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: 0.43 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.49 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.52 C4orf30chromosome 4 open reading frame 30 (219717_at), score: -0.5 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.68 C5orf23chromosome 5 open reading frame 23 (219054_at), score: 0.52 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.55 CANXcalnexin (208853_s_at), score: -0.51 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.4 CARD10caspase recruitment domain family, member 10 (210026_s_at), score: -0.52 CASP1caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) (211368_s_at), score: -0.51 CBLL1Cas-Br-M (murine) ecotropic retroviral transforming sequence-like 1 (220018_at), score: -0.53 CD24CD24 molecule (266_s_at), score: -0.51 CD81CD81 molecule (200675_at), score: 0.36 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: -0.51 CDK5R1cyclin-dependent kinase 5, regulatory subunit 1 (p35) (204995_at), score: 0.38 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.38 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.62 CFHR2complement factor H-related 2 (206910_x_at), score: -0.53 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 0.38 CH25Hcholesterol 25-hydroxylase (206932_at), score: 0.37 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: -0.57 CHPcalcium binding protein P22 (214665_s_at), score: 0.39 CHPFchondroitin polymerizing factor (202175_at), score: 0.39 CKBcreatine kinase, brain (200884_at), score: 0.38 CLINT1clathrin interactor 1 (201768_s_at), score: -0.61 CMTM6CKLF-like MARVEL transmembrane domain containing 6 (217947_at), score: -0.52 CNP2',3'-cyclic nucleotide 3' phosphodiesterase (208912_s_at), score: 0.42 COL11A1collagen, type XI, alpha 1 (37892_at), score: 0.4 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.5 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.44 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.59 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: 0.46 CRATcarnitine acetyltransferase (209522_s_at), score: 0.44 CREBL2cAMP responsive element binding protein-like 2 (201990_s_at), score: 0.39 CRIP2cysteine-rich protein 2 (208978_at), score: 0.38 CRTAPcartilage associated protein (201380_at), score: 0.52 CRY1cryptochrome 1 (photolyase-like) (209674_at), score: -0.51 CRYABcrystallin, alpha B (209283_at), score: 0.62 CTBP1C-terminal binding protein 1 (213979_s_at), score: -0.61 CTNNBL1catenin, beta like 1 (221021_s_at), score: -0.52 CTTNcortactin (201059_at), score: 0.37 CUX1cut-like homeobox 1 (214743_at), score: 0.41 CXorf15chromosome X open reading frame 15 (219969_at), score: -0.5 CYBAcytochrome b-245, alpha polypeptide (203028_s_at), score: 0.48 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.38 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.88 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: -0.57 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.71 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: 0.43 DGCR8DiGeorge syndrome critical region gene 8 (218650_at), score: 0.38 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.57 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (209178_at), score: -0.53 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.61 DIO2deiodinase, iodothyronine, type II (203700_s_at), score: -0.57 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: 0.42 DND1dead end homolog 1 (zebrafish) (57739_at), score: -0.59 DPYSL4dihydropyrimidinase-like 4 (205493_s_at), score: 0.4 DSPdesmoplakin (200606_at), score: 0.36 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.51 E4F1E4F transcription factor 1 (218524_at), score: -0.5 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.43 ECSITECSIT homolog (Drosophila) (218225_at), score: -0.57 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: 0.47 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: -0.54 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: -0.57 EPB41L3erythrocyte membrane protein band 4.1-like 3 (206710_s_at), score: 0.4 EPHB2EPH receptor B2 (209589_s_at), score: -0.63 EPS8L2EPS8-like 2 (218180_s_at), score: 0.37 ERO1LERO1-like (S. cerevisiae) (218498_s_at), score: 0.44 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: -0.55 EWSR1Ewing sarcoma breakpoint region 1 (210011_s_at), score: -0.5 FAF2Fas associated factor family member 2 (212106_at), score: -0.52 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: 0.59 FAM155Afamily with sequence similarity 155, member A (214825_at), score: 0.39 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.71 FAM60Afamily with sequence similarity 60, member A (220147_s_at), score: -0.5 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.37 FARP1FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) (201911_s_at), score: 0.36 FASFas (TNF receptor superfamily, member 6) (204780_s_at), score: 0.47 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.59 FBN2fibrillin 2 (203184_at), score: 0.49 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: 0.46 FHOD3formin homology 2 domain containing 3 (218980_at), score: 0.38 FKBP4FK506 binding protein 4, 59kDa (200894_s_at), score: -0.6 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.68 FLJ11151hypothetical protein FLJ11151 (218610_s_at), score: 0.36 FOLR3folate receptor 3 (gamma) (206371_at), score: 0.39 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: -0.58 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.49 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: -0.58 GAGE4G antigen 4 (207086_x_at), score: -0.9 GAGE6G antigen 6 (208155_x_at), score: -0.86 GAS6growth arrest-specific 6 (202177_at), score: 0.46 GATA3GATA binding protein 3 (209604_s_at), score: -0.76 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (202269_x_at), score: -0.58 GHRgrowth hormone receptor (205498_at), score: 0.44 GLE1GLE1 RNA export mediator homolog (yeast) (206920_s_at), score: -0.59 GLSglutaminase (203159_at), score: 0.49 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.36 GLTPglycolipid transfer protein (219267_at), score: 0.57 GMDSGDP-mannose 4,6-dehydratase (204875_s_at), score: -0.59 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: 0.41 GNL3guanine nucleotide binding protein-like 3 (nucleolar) (217850_at), score: -0.52 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: 0.35 GRK6G protein-coupled receptor kinase 6 (211543_s_at), score: -0.49 GSPT2G1 to S phase transition 2 (205541_s_at), score: 0.46 GTF2H4general transcription factor IIH, polypeptide 4, 52kDa (203577_at), score: -0.52 GTF2Igeneral transcription factor II, i (210892_s_at), score: -0.5 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: 0.49 H1F0H1 histone family, member 0 (208886_at), score: -0.55 H2AFYH2A histone family, member Y (214500_at), score: -0.5 HAS2hyaluronan synthase 2 (206432_at), score: 0.57 HEPHhephaestin (203903_s_at), score: 0.4 HERC5hect domain and RLD 5 (219863_at), score: -0.69 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.54 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: -0.57 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: -0.57 HIRIP3HIRA interacting protein 3 (204504_s_at), score: -0.49 HIST1H4Chistone cluster 1, H4c (205967_at), score: 0.58 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: 0.45 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.52 HLA-DPA1major histocompatibility complex, class II, DP alpha 1 (211990_at), score: -0.5 HNRNPA1heterogeneous nuclear ribonucleoprotein A1 (214280_x_at), score: -0.5 HNRNPCheterogeneous nuclear ribonucleoprotein C (C1/C2) (200751_s_at), score: -0.58 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: -0.61 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (200593_s_at), score: -0.5 HOXA10homeobox A10 (213150_at), score: 0.37 HRASLSHRAS-like suppressor (219983_at), score: -0.5 HSP90AB1heat shock protein 90kDa alpha (cytosolic), class B member 1 (214359_s_at), score: -0.5 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: -0.53 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.6 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.53 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.36 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: 0.68 HSPG2heparan sulfate proteoglycan 2 (201655_s_at), score: 0.37 IDEinsulin-degrading enzyme (203327_at), score: 0.36 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: -0.61 IFI44interferon-induced protein 44 (214453_s_at), score: -0.49 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: -0.57 IFRD1interferon-related developmental regulator 1 (202147_s_at), score: -0.55 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (203819_s_at), score: -0.49 IGFBP3insulin-like growth factor binding protein 3 (212143_s_at), score: 0.58 IKIK cytokine, down-regulator of HLA II (200066_at), score: -0.51 IKZF5IKAROS family zinc finger 5 (Pegasus) (220086_at), score: -0.53 IL21Rinterleukin 21 receptor (219971_at), score: 0.41 IL7interleukin 7 (206693_at), score: -0.5 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.37 INHBBinhibin, beta B (205258_at), score: 0.37 IPO8importin 8 (205701_at), score: -0.63 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.54 IRX5iroquois homeobox 5 (210239_at), score: 0.43 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.57 ITGBL1integrin, beta-like 1 (with EGF-like repeat domains) (214927_at), score: 0.44 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: -0.5 JARID1Djumonji, AT rich interactive domain 1D (206700_s_at), score: 0.41 JMJD4jumonji domain containing 4 (218560_s_at), score: 0.37 KIAA0355KIAA0355 (203288_at), score: 0.44 KIAA0495KIAA0495 (213340_s_at), score: 0.61 KIAA1539KIAA1539 (207765_s_at), score: 0.39 KIF13Bkinesin family member 13B (202962_at), score: -0.54 KIF3Akinesin family member 3A (213623_at), score: -0.58 KIFC1kinesin family member C1 (209680_s_at), score: -0.51 KLF4Kruppel-like factor 4 (gut) (221841_s_at), score: 0.39 KRIT1KRIT1, ankyrin repeat containing (34031_i_at), score: -0.53 KRT14keratin 14 (209351_at), score: 0.5 KRT19keratin 19 (201650_at), score: 0.52 LAMB3laminin, beta 3 (209270_at), score: -0.61 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.42 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: 0.41 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.55 LIMK2LIM domain kinase 2 (202193_at), score: 0.39 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: 0.42 LMF1lipase maturation factor 1 (219135_s_at), score: 0.38 LOC151162hypothetical LOC151162 (212098_at), score: 0.41 LOC388796hypothetical LOC388796 (65588_at), score: -0.59 LOC399491LOC399491 protein (214035_x_at), score: -0.51 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.59 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: -0.58 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: -0.57 LRP3low density lipoprotein receptor-related protein 3 (204381_at), score: 0.43 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.47 LRRFIP1leucine rich repeat (in FLII) interacting protein 1 (201861_s_at), score: 0.53 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.53 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.43 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: -0.65 MAD1L1MAD1 mitotic arrest deficient-like 1 (yeast) (204857_at), score: -0.5 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: 0.41 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: -0.54 MALLmal, T-cell differentiation protein-like (209373_at), score: 0.46 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: 0.59 MAP3K3mitogen-activated protein kinase kinase kinase 3 (203514_at), score: 0.42 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.5 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.79 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: -0.54 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: 0.5 MED6mediator complex subunit 6 (207079_s_at), score: -0.61 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.48 MFAP5microfibrillar associated protein 5 (213764_s_at), score: 0.6 MFGE8milk fat globule-EGF factor 8 protein (210605_s_at), score: 0.37 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: -0.5 MITFmicrophthalmia-associated transcription factor (207233_s_at), score: -0.59 MREGmelanoregulin (219648_at), score: 0.43 MRFAP1L1Morf4 family associated protein 1-like 1 (212199_at), score: -0.57 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: -0.56 MT1Gmetallothionein 1G (204745_x_at), score: 0.4 MT1Mmetallothionein 1M (217546_at), score: 0.55 MT1P3metallothionein 1 pseudogene 3 (221953_s_at), score: 0.51 MT1Xmetallothionein 1X (204326_x_at), score: 0.43 MXRA8matrix-remodelling associated 8 (213422_s_at), score: 0.43 MYO10myosin X (201976_s_at), score: 0.47 MYO1Cmyosin IC (214656_x_at), score: 0.7 MYO1Dmyosin ID (212338_at), score: 0.39 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: -0.63 NBASneuroblastoma amplified sequence (202926_at), score: 0.41 NBPF14neuroblastoma breakpoint family, member 14 (201104_x_at), score: -0.53 NCALDneurocalcin delta (211685_s_at), score: 0.39 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.49 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: 0.46 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: -0.73 NDNnecdin homolog (mouse) (209550_at), score: 0.48 NEFLneurofilament, light polypeptide (221805_at), score: -0.67 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: 0.44 NFYBnuclear transcription factor Y, beta (218129_s_at), score: -0.51 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: -0.58 NMUneuromedin U (206023_at), score: -0.56 NOC2Lnucleolar complex associated 2 homolog (S. cerevisiae) (202115_s_at), score: -0.54 NOL12nucleolar protein 12 (219324_at), score: -0.55 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: -0.63 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.42 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: 0.39 NR2F6nuclear receptor subfamily 2, group F, member 6 (209262_s_at), score: 0.37 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.62 NTF3neurotrophin 3 (206706_at), score: 0.41 NUP50nucleoporin 50kDa (218295_s_at), score: -0.54 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.99 OAZ3ornithine decarboxylase antizyme 3 (222075_s_at), score: 0.38 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.42 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.5 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: 0.36 PAOXpolyamine oxidase (exo-N4-amino) (50400_at), score: 0.36 PARP4poly (ADP-ribose) polymerase family, member 4 (202239_at), score: -0.53 PBLDphenazine biosynthesis-like protein domain containing (219543_at), score: 0.38 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.43 PCBP4poly(rC) binding protein 4 (209361_s_at), score: 0.39 PCOLCE2procollagen C-endopeptidase enhancer 2 (219295_s_at), score: 0.37 PDAP1PDGFA associated protein 1 (202290_at), score: -0.53 PDE10Aphosphodiesterase 10A (205501_at), score: -0.53 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: 0.56 PGRMC2progesterone receptor membrane component 2 (213227_at), score: 0.38 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.51 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.69 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.44 PLXNA2plexin A2 (213030_s_at), score: 0.38 PODXLpodocalyxin-like (201578_at), score: 0.53 PORCNporcupine homolog (Drosophila) (219483_s_at), score: 0.36 PPCDCphosphopantothenoylcysteine decarboxylase (219066_at), score: -0.51 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: -0.59 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: 0.47 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: 0.37 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: 0.36 PQBP1polyglutamine binding protein 1 (214527_s_at), score: -0.52 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.36 PRPF38BPRP38 pre-mRNA processing factor 38 (yeast) domain containing B (218040_at), score: -0.51 PRPF4PRP4 pre-mRNA processing factor 4 homolog (yeast) (209162_s_at), score: -0.52 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: 0.64 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.62 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: -0.51 PSMC3proteasome (prosome, macropain) 26S subunit, ATPase, 3 (201267_s_at), score: -0.49 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (202353_s_at), score: 0.36 PSMD13proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 (201233_at), score: -0.53 PTGER3prostaglandin E receptor 3 (subtype EP3) (213933_at), score: 0.41 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.36 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.45 PTNpleiotrophin (209466_x_at), score: -0.49 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: 0.56 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.42 PXDNperoxidasin homolog (Drosophila) (212013_at), score: 0.48 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.42 R3HDM2R3H domain containing 2 (203831_at), score: 0.38 RAB40BRAB40B, member RAS oncogene family (204547_at), score: 0.46 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: -0.66 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: 0.47 RABGEF1RAB guanine nucleotide exchange factor (GEF) 1 (218310_at), score: 0.38 RAD23BRAD23 homolog B (S. cerevisiae) (201223_s_at), score: 0.68 RASIP1Ras interacting protein 1 (220027_s_at), score: 0.39 RBM12BRNA binding motif protein 12B (51228_at), score: -0.49 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: 0.53 RBM5RNA binding motif protein 5 (201394_s_at), score: -0.51 RCBTB2regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 (204759_at), score: 0.42 RDXradixin (204969_s_at), score: -0.54 REEP1receptor accessory protein 1 (204364_s_at), score: -0.5 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (212968_at), score: 0.36 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: -0.56 RHBDF2rhomboid 5 homolog 2 (Drosophila) (219202_at), score: 0.37 RHODras homolog gene family, member D (31846_at), score: 0.44 RIN2Ras and Rab interactor 2 (209684_at), score: 0.4 RNF19Aring finger protein 19A (220483_s_at), score: -0.56 RORARAR-related orphan receptor A (210426_x_at), score: 0.38 RP6-213H19.1serine/threonine protein kinase MST4 (218499_at), score: -0.53 RPL31ribosomal protein L31 (221593_s_at), score: 0.4 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: -0.59 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.42 RTN3reticulon 3 (219549_s_at), score: 0.37 SAFB2scaffold attachment factor B2 (32099_at), score: -0.6 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.42 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: -0.62 SC65synaptonemal complex protein SC65 (204078_at), score: 0.35 SCG5secretogranin V (7B2 protein) (203889_at), score: 0.5 SDC4syndecan 4 (202071_at), score: 0.36 SEC16ASEC16 homolog A (S. cerevisiae) (215696_s_at), score: 0.36 SEC61A1Sec61 alpha 1 subunit (S. cerevisiae) (217716_s_at), score: 0.36 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: -0.5 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.38 SERTAD2SERTA domain containing 2 (202657_s_at), score: 0.37 SF3B1splicing factor 3b, subunit 1, 155kDa (201070_x_at), score: -0.52 SF4splicing factor 4 (215004_s_at), score: -0.55 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: -0.51 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.49 SHANK2SH3 and multiple ankyrin repeat domains 2 (213307_at), score: 0.38 SHFM1split hand/foot malformation (ectrodactyly) type 1 (202276_at), score: -0.51 SIDT2SID1 transmembrane family, member 2 (56256_at), score: 0.46 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: 0.41 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: 0.55 SLC1A1solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 (213664_at), score: 0.44 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: 0.37 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: 0.39 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: 0.38 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.54 SLC35A3solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 (206770_s_at), score: 0.44 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: 0.59 SLC7A1solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 (212295_s_at), score: 0.4 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: -0.57 SMAD5SMAD family member 5 (205187_at), score: -0.53 SMAD7SMAD family member 7 (204790_at), score: 0.39 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.67 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.63 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: 0.38 SNAPC1small nuclear RNA activating complex, polypeptide 1, 43kDa (205443_at), score: -0.5 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.82 SPARCsecreted protein, acidic, cysteine-rich (osteonectin) (212667_at), score: 0.46 SPINT2serine peptidase inhibitor, Kunitz type, 2 (210715_s_at), score: 0.44 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: -0.55 SSNA1Sjogren syndrome nuclear autoantigen 1 (210378_s_at), score: -0.52 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.6 SSRP1structure specific recognition protein 1 (200956_s_at), score: -0.5 STX16syntaxin 16 (221499_s_at), score: -0.57 SUPT16Hsuppressor of Ty 16 homolog (S. cerevisiae) (217815_at), score: -0.53 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.68 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.48 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.59 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.52 TBX5T-box 5 (207155_at), score: 0.35 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.82 TEKTEK tyrosine kinase, endothelial (206702_at), score: 0.42 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: -0.6 TESK1testis-specific kinase 1 (204106_at), score: 0.39 TGFAtransforming growth factor, alpha (205016_at), score: 0.38 TGIF1TGFB-induced factor homeobox 1 (203313_s_at), score: -0.55 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.55 TMC7transmembrane channel-like 7 (220021_at), score: 0.4 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: 0.37 TMEM204transmembrane protein 204 (219315_s_at), score: 0.57 TMEM47transmembrane protein 47 (209656_s_at), score: 0.49 TMEM51transmembrane protein 51 (218815_s_at), score: -0.62 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.43 TMEM87Atransmembrane protein 87A (212204_at), score: 0.35 TMEM97transmembrane protein 97 (212279_at), score: -0.53 TMOD1tropomodulin 1 (203661_s_at), score: -0.66 TMX1thioredoxin-related transmembrane protein 1 (208097_s_at), score: -0.5 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: 0.4 TNFRSF21tumor necrosis factor receptor superfamily, member 21 (214581_x_at), score: -0.49 TNS1tensin 1 (221748_s_at), score: 0.52 TOB1transducer of ERBB2, 1 (202704_at), score: 0.51 TRIM16tripartite motif-containing 16 (204341_at), score: -0.6 TRIM8tripartite motif-containing 8 (221012_s_at), score: 0.39 TSNtranslin (201504_s_at), score: -0.56 TSPAN13tetraspanin 13 (217979_at), score: 0.52 TSPYL1TSPY-like 1 (221493_at), score: 0.46 TSPYL5TSPY-like 5 (213122_at), score: 0.55 TTTY15testis-specific transcript, Y-linked 15 (214983_at), score: 0.43 TYRP1tyrosinase-related protein 1 (205694_at), score: -0.51 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: 0.62 UBE2D4ubiquitin-conjugating enzyme E2D 4 (putative) (218837_s_at), score: 0.44 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: -0.63 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.54 UBL3ubiquitin-like 3 (201535_at), score: 0.56 UBTFupstream binding transcription factor, RNA polymerase I (214881_s_at), score: -0.53 UCHL1ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (201387_s_at), score: 0.49 UGCGUDP-glucose ceramide glucosyltransferase (204881_s_at), score: -0.54 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.83 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.86 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.85 UROSuroporphyrinogen III synthase (203031_s_at), score: 0.48 USP18ubiquitin specific peptidase 18 (219211_at), score: -0.6 USP25ubiquitin specific peptidase 25 (220419_s_at), score: 0.46 UXS1UDP-glucuronate decarboxylase 1 (219675_s_at), score: 0.4 VEGFCvascular endothelial growth factor C (209946_at), score: 0.39 VKORC1vitamin K epoxide reductase complex, subunit 1 (217949_s_at), score: 0.39 VLDLRvery low density lipoprotein receptor (209822_s_at), score: 0.47 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.55 WDR74WD repeat domain 74 (221712_s_at), score: -0.5 WDR91WD repeat domain 91 (218971_s_at), score: -0.56 WISP1WNT1 inducible signaling pathway protein 1 (206796_at), score: -0.52 WNT5Bwingless-type MMTV integration site family, member 5B (221029_s_at), score: 0.39 WT1Wilms tumor 1 (206067_s_at), score: -0.6 WWC3WWC family member 3 (219520_s_at), score: 0.38 XYLT1xylosyltransferase I (213725_x_at), score: 0.36 YIPF2Yip1 domain family, member 2 (65086_at), score: 0.37 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: 0.46 ZNF224zinc finger protein 224 (216983_s_at), score: -0.6 ZNF227zinc finger protein 227 (217403_s_at), score: -0.59 ZNF281zinc finger protein 281 (218401_s_at), score: 0.52 ZNF331zinc finger protein 331 (219228_at), score: -0.52 ZNF415zinc finger protein 415 (205514_at), score: 0.49 ZNF562zinc finger protein 562 (219163_at), score: 0.37 ZNF654zinc finger protein 654 (219239_s_at), score: 0.45 ZNF804Azinc finger protein 804A (215767_at), score: -0.52
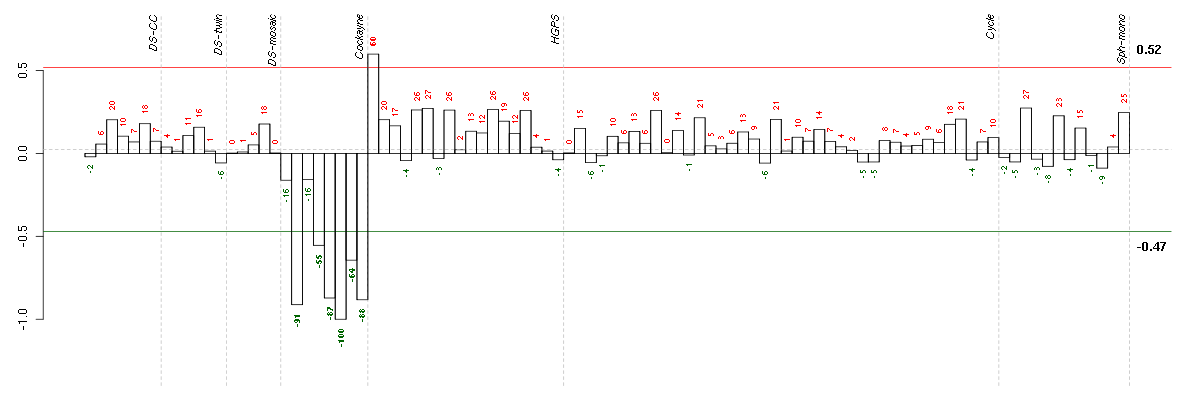
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |