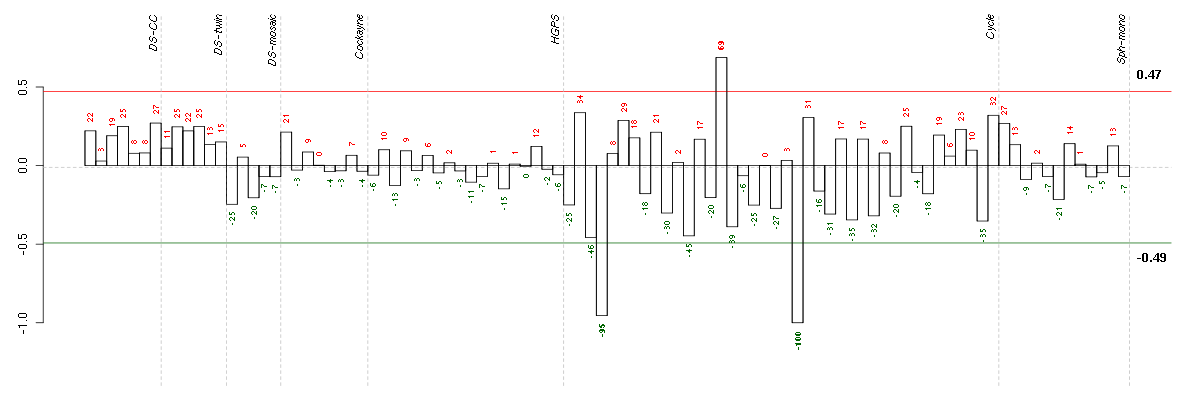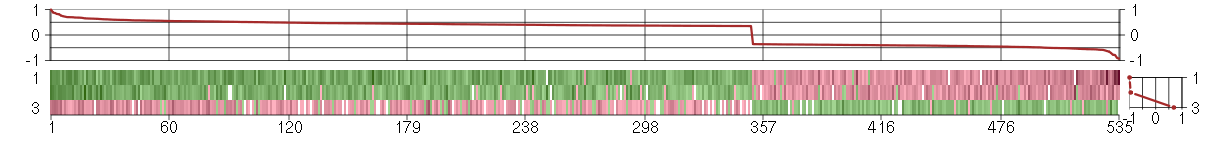

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
A1CFAPOBEC1 complementation factor (220951_s_at), score: 0.56 ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: -0.4 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.38 ABOABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (216716_at), score: 0.44 ACAD10acyl-Coenzyme A dehydrogenase family, member 10 (219986_s_at), score: 0.46 ACADSacyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain (202366_at), score: 0.51 ACADSBacyl-Coenzyme A dehydrogenase, short/branched chain (205355_at), score: 0.37 ACVR1Bactivin A receptor, type IB (213198_at), score: 0.42 ACVR2Aactivin A receptor, type IIA (205327_s_at), score: 0.4 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: 0.52 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.44 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.52 ADORA1adenosine A1 receptor (216220_s_at), score: 0.44 AGAP7ArfGAP with GTPase domain, ankyrin repeat and PH domain 7 (221971_x_at), score: 0.41 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: 0.39 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: 0.37 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.5 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: 0.42 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.46 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: -0.44 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: 0.41 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: -0.43 ANO1anoctamin 1, calcium activated chloride channel (218804_at), score: -0.37 APBA1amyloid beta (A4) precursor protein-binding, family A, member 1 (206679_at), score: 0.37 APBB1IPamyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein (219994_at), score: -0.45 APOBEC3Aapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A (210873_x_at), score: 0.45 APOC4apolipoprotein C-IV (206738_at), score: 0.53 APOL2apolipoprotein L, 2 (221653_x_at), score: 0.44 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: -0.39 ARHGEF10LRho guanine nucleotide exchange factor (GEF) 10-like (221656_s_at), score: 0.42 ARHGEF15Rho guanine nucleotide exchange factor (GEF) 15 (217348_x_at), score: 0.37 ARL15ADP-ribosylation factor-like 15 (219842_at), score: -0.36 ARSAarylsulfatase A (204443_at), score: -0.45 ASB13ankyrin repeat and SOCS box-containing 13 (218862_at), score: 0.56 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: 0.38 ATF5activating transcription factor 5 (204999_s_at), score: -0.4 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.35 ATP13A2ATPase type 13A2 (218608_at), score: -0.39 ATP6V1G2ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 (214762_at), score: 0.36 ATP8A2ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 (219660_s_at), score: -0.42 ATXN1ataxin 1 (203232_s_at), score: 0.39 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: 0.44 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (221234_s_at), score: -0.53 BAGEB melanoma antigen (207712_at), score: -0.51 BAIAP2BAI1-associated protein 2 (205294_at), score: -0.36 BAIAP3BAI1-associated protein 3 (216356_x_at), score: 0.59 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.4 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.38 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.37 BCANbrevican (91920_at), score: 0.5 BCL6B-cell CLL/lymphoma 6 (203140_at), score: 0.36 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.38 BTNL3butyrophilin-like 3 (217207_s_at), score: 0.56 C10orf110chromosome 10 open reading frame 110 (220703_at), score: 0.54 C10orf81chromosome 10 open reading frame 81 (219857_at), score: -0.93 C11orf30chromosome 11 open reading frame 30 (219012_s_at), score: -0.46 C16orf53chromosome 16 open reading frame 53 (218300_at), score: -0.43 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.65 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: -0.41 C1orf61chromosome 1 open reading frame 61 (205103_at), score: 0.51 C1orf69chromosome 1 open reading frame 69 (215490_at), score: 0.58 C1orf89chromosome 1 open reading frame 89 (220963_s_at), score: 0.37 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.36 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: 0.38 C4Acomplement component 4A (Rodgers blood group) (214428_x_at), score: 0.68 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.52 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.51 C8orf17chromosome 8 open reading frame 17 (208266_at), score: 0.46 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: 0.4 C8orf60chromosome 8 open reading frame 60 (220712_at), score: 0.67 C8orf84chromosome 8 open reading frame 84 (214725_at), score: -0.47 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.43 C9orf61chromosome 9 open reading frame 61 (213900_at), score: -0.37 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: 0.45 CALB1calbindin 1, 28kDa (205626_s_at), score: 0.64 CALML4calmodulin-like 4 (64408_s_at), score: 0.44 CAMK1Gcalcium/calmodulin-dependent protein kinase IG (217128_s_at), score: 0.43 CAPZA1capping protein (actin filament) muscle Z-line, alpha 1 (208374_s_at), score: 0.38 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: 0.35 CCBP2chemokine binding protein 2 (206887_at), score: 0.37 CCNL1cyclin L1 (220046_s_at), score: 0.44 CD34CD34 molecule (209543_s_at), score: 0.38 CD53CD53 molecule (203416_at), score: 0.57 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.82 CD80CD80 molecule (207176_s_at), score: -0.44 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: 0.37 CDC23cell division cycle 23 homolog (S. cerevisiae) (202892_at), score: 0.41 CDH19cadherin 19, type 2 (206898_at), score: 0.4 CDH4cadherin 4, type 1, R-cadherin (retinal) (220227_at), score: 0.36 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: -0.42 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.36 CHPcalcium binding protein P22 (214665_s_at), score: 0.44 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: 0.48 CLEC2DC-type lectin domain family 2, member D (220132_s_at), score: 0.39 CLEC7AC-type lectin domain family 7, member A (221698_s_at), score: 0.5 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: 0.36 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: 0.37 CNNM1cyclin M1 (220166_at), score: 0.36 CNNM2cyclin M2 (206818_s_at), score: 0.44 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.39 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.74 COL6A1collagen, type VI, alpha 1 (212940_at), score: -0.36 COL9A2collagen, type IX, alpha 2 (213622_at), score: 0.65 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.43 COX16COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) (217645_at), score: 0.4 COX5Bcytochrome c oxidase subunit Vb (213736_at), score: 0.36 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: 0.48 CRCPCGRP receptor component (203898_at), score: 0.35 CRKRSCdc2-related kinase, arginine/serine-rich (219226_at), score: -0.58 CRYBG3beta-gamma crystallin domain containing 3 (214030_at), score: 0.39 CSF2RBcolony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) (205159_at), score: -0.41 CSN2casein beta (207951_at), score: -0.42 CTNNB1catenin (cadherin-associated protein), beta 1, 88kDa (201533_at), score: 0.36 CTRLchymotrypsin-like (214377_s_at), score: 0.48 CTTNcortactin (201059_at), score: 0.48 CXorf56chromosome X open reading frame 56 (219805_at), score: 0.37 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.46 CYTH4cytohesin 4 (219183_s_at), score: 0.44 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.35 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: -0.4 DENND4BDENN/MADD domain containing 4B (202860_at), score: -0.37 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: 0.54 DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (206752_s_at), score: -0.52 DGKEdiacylglycerol kinase, epsilon 64kDa (207518_at), score: -0.43 DGKIdiacylglycerol kinase, iota (206806_at), score: -0.36 DHRS2dehydrogenase/reductase (SDR family) member 2 (214079_at), score: -0.53 DISC1disrupted in schizophrenia 1 (206090_s_at), score: 0.37 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: 0.4 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: 0.48 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: -0.39 DOK4docking protein 4 (209691_s_at), score: -0.44 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.86 DUSP8dual specificity phosphatase 8 (206374_at), score: 0.55 DUSP9dual specificity phosphatase 9 (205777_at), score: 0.4 EDARectodysplasin A receptor (220048_at), score: 0.43 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.35 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: -0.56 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: -0.47 ELA3Aelastase 3A, pancreatic (211738_x_at), score: 0.44 ELAC1elaC homolog 1 (E. coli) (219325_s_at), score: -0.41 ENO3enolase 3 (beta, muscle) (204483_at), score: 0.49 ENOX2ecto-NOX disulfide-thiol exchanger 2 (32042_at), score: 0.41 EPAGearly lymphoid activation protein (217050_at), score: 0.48 EPB41L5erythrocyte membrane protein band 4.1 like 5 (220977_x_at), score: 0.51 EPHA2EPH receptor A2 (203499_at), score: -0.36 EPHB3EPH receptor B3 (1438_at), score: 0.38 ERFEts2 repressor factor (203643_at), score: 0.36 ERMAPerythroblast membrane-associated protein (Scianna blood group) (219905_at), score: 0.4 ETV7ets variant 7 (221680_s_at), score: 0.57 EVLEnah/Vasp-like (217838_s_at), score: -0.43 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: 0.57 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.62 FBRSfibrosin (218255_s_at), score: 0.68 FBXL6F-box and leucine-rich repeat protein 6 (219189_at), score: -0.46 FBXO11F-box protein 11 (203255_at), score: 0.44 FBXO2F-box protein 2 (219305_x_at), score: 0.7 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: 0.61 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: 0.48 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: -0.36 FETUBfetuin B (214417_s_at), score: 0.61 FGF12fibroblast growth factor 12 (207501_s_at), score: 0.39 FGF13fibroblast growth factor 13 (205110_s_at), score: -0.41 FGL1fibrinogen-like 1 (205305_at), score: 0.82 FHOD1formin homology 2 domain containing 1 (218530_at), score: -0.41 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.57 FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.57 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: 0.52 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: -0.56 FNDC4fibronectin type III domain containing 4 (218843_at), score: -0.4 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: -0.45 FOXK2forkhead box K2 (203064_s_at), score: -0.37 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: -0.38 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.45 FUT4fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) (209893_s_at), score: 0.46 GAB1GRB2-associated binding protein 1 (207112_s_at), score: -0.44 GABRR1gamma-aminobutyric acid (GABA) receptor, rho 1 (206525_at), score: 0.48 GAKcyclin G associated kinase (202281_at), score: -0.37 GANgigaxonin (220124_at), score: -0.58 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.52 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.38 GDF9growth differentiation factor 9 (221314_at), score: 0.49 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: -0.37 GGT1gamma-glutamyltransferase 1 (209919_x_at), score: 0.58 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: 0.45 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: 0.48 GNG4guanine nucleotide binding protein (G protein), gamma 4 (205184_at), score: 0.49 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: 0.42 GOLGA9Pgolgi autoantigen, golgin subfamily a, 9 pseudogene (213737_x_at), score: 0.35 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: 0.48 GP6glycoprotein VI (platelet) (220336_s_at), score: -0.63 GPATCH4G patch domain containing 4 (220596_at), score: 0.46 GPC4glypican 4 (204983_s_at), score: 0.42 GPD2glycerol-3-phosphate dehydrogenase 2 (mitochondrial) (210007_s_at), score: -0.37 GPR144G protein-coupled receptor 144 (216289_at), score: 0.7 GPR32G protein-coupled receptor 32 (221469_at), score: 0.65 GRB10growth factor receptor-bound protein 10 (210999_s_at), score: 0.52 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.4 GSTA1glutathione S-transferase alpha 1 (215766_at), score: 0.37 HAB1B1 for mucin (215778_x_at), score: 0.72 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.51 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.38 HECW1HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 (215584_at), score: 0.46 HERC3hect domain and RLD 3 (206183_s_at), score: -0.38 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.42 HIST1H2ALhistone cluster 1, H2al (214554_at), score: 0.4 HIST1H3Hhistone cluster 1, H3h (206110_at), score: 0.43 HIST1H4Dhistone cluster 1, H4d (208076_at), score: 0.38 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.52 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: 0.38 HLCSholocarboxylase synthetase (biotin-(proprionyl-Coenzyme A-carboxylase (ATP-hydrolysing)) ligase) (209399_at), score: 0.38 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.39 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.37 HOXB5homeobox B5 (205601_s_at), score: 0.37 HPNhepsin (204934_s_at), score: 0.47 HPS4Hermansky-Pudlak syndrome 4 (54037_at), score: 0.36 HS3ST2heparan sulfate (glucosamine) 3-O-sulfotransferase 2 (219697_at), score: 0.42 HTR1E5-hydroxytryptamine (serotonin) receptor 1E (207404_s_at), score: 0.47 HYIhydroxypyruvate isomerase homolog (E. coli) (221435_x_at), score: -0.36 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.57 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.54 IL15RAinterleukin 15 receptor, alpha (207375_s_at), score: 0.48 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: 0.38 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.42 ING4inhibitor of growth family, member 4 (48825_at), score: 0.37 INPP4Binositol polyphosphate-4-phosphatase, type II, 105kDa (205376_at), score: -0.39 IRF2interferon regulatory factor 2 (203275_at), score: -0.41 ISOC2isochorismatase domain containing 2 (218893_at), score: 0.54 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.88 JAG2jagged 2 (32137_at), score: 0.42 KCNAB1potassium voltage-gated channel, shaker-related subfamily, beta member 1 (210078_s_at), score: -0.45 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.43 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: 0.37 KIAA0146KIAA0146 (212523_s_at), score: -0.37 KIAA0355KIAA0355 (203288_at), score: -0.44 KIAA0652KIAA0652 (203363_s_at), score: -0.38 KIAA1467KIAA1467 (213234_at), score: 0.37 KLF12Kruppel-like factor 12 (208467_at), score: -0.42 KRT14keratin 14 (209351_at), score: -0.47 KRT24keratin 24 (220267_at), score: 0.51 KRT33Akeratin 33A (208483_x_at), score: -0.41 KRT9keratin 9 (208188_at), score: 0.42 KRTAP1-1keratin associated protein 1-1 (220976_s_at), score: -0.45 KSR1kinase suppressor of ras 1 (213769_at), score: 0.36 LEPREL2leprecan-like 2 (204854_at), score: -0.38 LGALS14lectin, galactoside-binding, soluble, 14 (220158_at), score: 0.37 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.61 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.53 LMF2lipase maturation factor 2 (212682_s_at), score: -0.4 LOC100129064hypothetical LOC100129064 (220764_at), score: -0.41 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: 0.46 LOC100133503similar to LRRC37A3 protein (220219_s_at), score: 0.36 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: 0.4 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.62 LOC149501similar to keratin 8 (216821_at), score: 0.4 LOC150759hypothetical protein LOC150759 (213703_at), score: 0.47 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.41 LOC391132similar to hCG2041276 (216177_at), score: 0.5 LOC80054hypothetical LOC80054 (220465_at), score: 0.59 LPHN1latrophilin 1 (203488_at), score: 0.4 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.56 LRRC37A2leucine rich repeat containing 37, member A2 (221740_x_at), score: 0.65 LY6Hlymphocyte antigen 6 complex, locus H (206773_at), score: 0.42 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: 0.47 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: -0.37 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.37 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.56 MAPK8mitogen-activated protein kinase 8 (210671_x_at), score: 0.46 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.43 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: -0.4 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: 0.39 MATN3matrilin 3 (206091_at), score: -0.9 MCATmalonyl CoA:ACP acyltransferase (mitochondrial) (213132_s_at), score: -0.48 MCM9minichromosome maintenance complex component 9 (219673_at), score: 0.37 MEN1multiple endocrine neoplasia I (202645_s_at), score: -0.42 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.64 MEP1Bmeprin A, beta (207251_at), score: -0.54 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.47 MID2midline 2 (208384_s_at), score: 0.41 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (203936_s_at), score: 0.35 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.45 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.46 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: 0.38 MTHFSDmethenyltetrahydrofolate synthetase domain containing (218879_s_at), score: -0.38 MTMR11myotubularin related protein 11 (205076_s_at), score: -0.41 MTMR8myotubularin related protein 8 (220537_at), score: -0.46 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: 0.47 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.54 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.55 MVKmevalonate kinase (36907_at), score: 0.39 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.36 MYO5Cmyosin VC (218966_at), score: 0.54 N4BP3Nedd4 binding protein 3 (214775_at), score: 0.53 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.68 NCAM1neural cell adhesion molecule 1 (212843_at), score: 0.4 NCAM2neural cell adhesion molecule 2 (205669_at), score: 0.42 NCKAP1NCK-associated protein 1 (207738_s_at), score: 0.37 NCLNnicalin homolog (zebrafish) (222206_s_at), score: -0.4 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.36 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.4 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: -0.4 NEURLneuralized homolog (Drosophila) (204889_s_at), score: 0.38 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.42 NHLH2nescient helix loop helix 2 (215228_at), score: -0.39 NINninein (GSK3B interacting protein) (219285_s_at), score: -0.5 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: 0.4 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.47 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: -0.37 NPEPL1aminopeptidase-like 1 (89476_r_at), score: 0.39 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: 0.39 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.45 NUPL2nucleoporin like 2 (204003_s_at), score: 0.5 NXF1nuclear RNA export factor 1 (208922_s_at), score: 0.36 NXPH3neurexophilin 3 (221991_at), score: 0.44 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.37 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: -0.51 OLAHoleoyl-ACP hydrolase (219975_x_at), score: 0.39 OPRL1opiate receptor-like 1 (206564_at), score: 0.53 OR1D2olfactory receptor, family 1, subfamily D, member 2 (221464_at), score: -0.4 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: 0.46 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: 0.59 PACS1phosphofurin acidic cluster sorting protein 1 (220557_s_at), score: -0.47 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.39 PAK2p21 protein (Cdc42/Rac)-activated kinase 2 (208878_s_at), score: -0.4 PARVBparvin, beta (37965_at), score: -0.44 PCDH24protocadherin 24 (220186_s_at), score: 0.49 PCMTD2protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 (212406_s_at), score: 0.36 PDE8Bphosphodiesterase 8B (213228_at), score: 0.53 PDK3pyruvate dehydrogenase kinase, isozyme 3 (221957_at), score: -0.55 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: 0.41 PEG3paternally expressed 3 (209242_at), score: 0.35 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.64 PGLYRP4peptidoglycan recognition protein 4 (220944_at), score: 0.35 PHC3polyhomeotic homolog 3 (Drosophila) (215521_at), score: 0.63 PHF2PHD finger protein 2 (212726_at), score: -0.39 PHF7PHD finger protein 7 (215622_x_at), score: 0.55 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: 0.38 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: -0.38 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (207105_s_at), score: -0.39 PIM1pim-1 oncogene (209193_at), score: 0.37 PIN1Lpeptidylprolyl cis/trans isomerase, NIMA-interacting 1-like (pseudogene) (207582_at), score: 0.56 PIP5K3phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (213111_at), score: 0.49 PISDphosphatidylserine decarboxylase (202392_s_at), score: -0.36 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.59 PKN1protein kinase N1 (202161_at), score: -0.46 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: -0.43 PLXNB1plexin B1 (215807_s_at), score: -0.37 POLG2polymerase (DNA directed), gamma 2, accessory subunit (205811_at), score: -0.44 POLHpolymerase (DNA directed), eta (219380_x_at), score: 0.43 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.41 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: 0.43 PPP1R10protein phosphatase 1, regulatory (inhibitor) subunit 10 (201702_s_at), score: -0.47 PPP1R9Aprotein phosphatase 1, regulatory (inhibitor) subunit 9A (221088_s_at), score: -0.78 PPP2R2Bprotein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (213849_s_at), score: 0.36 PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: 0.36 PPT2palmitoyl-protein thioesterase 2 (209490_s_at), score: -0.39 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: 0.37 PRKCBprotein kinase C, beta (209685_s_at), score: 0.42 PRKCDprotein kinase C, delta (202545_at), score: -0.46 PRLHprolactin releasing hormone (221443_x_at), score: 0.57 ProSAPiP1ProSAPiP1 protein (204447_at), score: 0.41 PRR14proline rich 14 (218714_at), score: -0.49 PTPN21protein tyrosine phosphatase, non-receptor type 21 (205438_at), score: -0.41 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 1 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: -0.38 PVT1Pvt1 oncogene (non-protein coding) (216249_at), score: 0.4 PXNpaxillin (211823_s_at), score: -0.56 PYGO1pygopus homolog 1 (Drosophila) (215517_at), score: 0.53 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: 0.57 RAB27BRAB27B, member RAS oncogene family (207018_s_at), score: -0.45 RAMP1receptor (G protein-coupled) activity modifying protein 1 (204916_at), score: 0.5 RANBP17RAN binding protein 17 (219661_at), score: 0.35 RASA3RAS p21 protein activator 3 (206220_s_at), score: -0.48 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.43 RBBP6retinoblastoma binding protein 6 (212781_at), score: 0.42 RBM12BRNA binding motif protein 12B (51228_at), score: -0.41 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: 0.38 RBM38RNA binding motif protein 38 (212430_at), score: 0.4 RCL1RNA terminal phosphate cyclase-like 1 (218544_s_at), score: 0.42 RFX3regulatory factor X, 3 (influences HLA class II expression) (207234_at), score: -0.78 RHEBRas homolog enriched in brain (213409_s_at), score: 0.46 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: 0.35 RICH2Rho-type GTPase-activating protein RICH2 (215232_at), score: 0.68 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: 0.44 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: -0.51 RLN1relaxin 1 (211753_s_at), score: 0.53 RMND5Brequired for meiotic nuclear division 5 homolog B (S. cerevisiae) (218262_at), score: -0.54 RNF220ring finger protein 220 (219988_s_at), score: -0.45 RNF40ring finger protein 40 (206845_s_at), score: -0.47 RORARAR-related orphan receptor A (210426_x_at), score: 0.49 RP3-377H14.5hypothetical LOC285830 (222279_at), score: 0.39 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.51 RPH3ALrabphilin 3A-like (without C2 domains) (221614_s_at), score: -0.41 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.56 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: 0.4 RPL27Aribosomal protein L27a (212044_s_at), score: 0.55 RPL38ribosomal protein L38 (202028_s_at), score: 0.56 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.45 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: -0.38 RPS11ribosomal protein S11 (213350_at), score: 0.44 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.4 RPS27Aribosomal protein S27a (217144_at), score: 0.37 RRP8ribosomal RNA processing 8, methyltransferase, homolog (yeast) (203171_s_at), score: -0.45 RYR3ryanodine receptor 3 (206306_at), score: 0.38 S100A14S100 calcium binding protein A14 (218677_at), score: 0.6 SAA4serum amyloid A4, constitutive (207096_at), score: 0.54 SASH3SAM and SH3 domain containing 3 (204923_at), score: 0.46 SBF1SET binding factor 1 (39835_at), score: -0.38 SCML2sex comb on midleg-like 2 (Drosophila) (206147_x_at), score: 0.38 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: 0.39 SEC14L5SEC14-like 5 (S. cerevisiae) (210169_at), score: 0.55 SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: -0.52 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: -0.36 SEPT5septin 5 (209767_s_at), score: 0.47 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: 0.45 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.54 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.49 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: -0.37 SKAP2src kinase associated phosphoprotein 2 (216899_s_at), score: -0.5 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: -0.39 SLC19A1solute carrier family 19 (folate transporter), member 1 (211576_s_at), score: 0.38 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.51 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: 0.51 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.36 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.51 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: 0.49 SLC4A5solute carrier family 4, sodium bicarbonate cotransporter, member 5 (204296_at), score: 0.5 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.37 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (201349_at), score: 0.52 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (212947_at), score: 0.45 SLCO2A1solute carrier organic anion transporter family, member 2A1 (204368_at), score: 0.54 SLTMSAFB-like, transcription modulator (217828_at), score: 0.36 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: -0.4 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.43 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: 0.41 SNRPEsmall nuclear ribonucleoprotein polypeptide E (215450_at), score: 0.36 SOX12SRY (sex determining region Y)-box 12 (204432_at), score: 0.36 SPATA1spermatogenesis associated 1 (221057_at), score: 0.7 SPATA6spermatogenesis associated 6 (220298_s_at), score: 0.57 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: 0.46 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: -0.48 SSX3synovial sarcoma, X breakpoint 3 (215881_x_at), score: 0.52 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.51 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.6 STIM1stromal interaction molecule 1 (202764_at), score: -0.7 SULT1A2sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2 (207122_x_at), score: 0.64 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.35 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: 0.36 SYT5synaptotagmin V (206161_s_at), score: 0.5 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.54 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.44 TAOK2TAO kinase 2 (204986_s_at), score: -0.58 TAPBPLTAP binding protein-like (218747_s_at), score: 0.37 TASP1taspase, threonine aspartase, 1 (219443_at), score: 0.46 TBX21T-box 21 (220684_at), score: 0.47 TBXA2Rthromboxane A2 receptor (336_at), score: -0.41 TDO2tryptophan 2,3-dioxygenase (205943_at), score: -0.43 TFDP1transcription factor Dp-1 (204147_s_at), score: -0.38 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: 0.6 THNSL1threonine synthase-like 1 (S. cerevisiae) (219800_s_at), score: -0.39 THRAthyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) (35846_at), score: 0.39 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.37 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: 0.36 TMC5transmembrane channel-like 5 (219580_s_at), score: 0.52 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.55 TMCO6transmembrane and coiled-coil domains 6 (221096_s_at), score: -0.37 TMEM35transmembrane protein 35 (219685_at), score: -0.55 TMEM40transmembrane protein 40 (219503_s_at), score: 0.53 TMEM92transmembrane protein 92 (216791_at), score: 0.56 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.62 TNFRSF14tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) (209354_at), score: 0.53 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: 0.35 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.54 TNK2tyrosine kinase, non-receptor, 2 (203839_s_at), score: -0.41 TRAPPC9trafficking protein particle complex 9 (221836_s_at), score: 0.44 TRIM3tripartite motif-containing 3 (204911_s_at), score: -0.36 TRPM3transient receptor potential cation channel, subfamily M, member 3 (220463_at), score: -0.39 TSHBthyroid stimulating hormone, beta (214529_at), score: -0.37 TSPAN12tetraspanin 12 (219274_at), score: -0.38 TSPAN32tetraspanin 32 (220558_x_at), score: 0.37 TSPAN9tetraspanin 9 (220968_s_at), score: 0.46 TSSC4tumor suppressing subtransferable candidate 4 (218612_s_at), score: -0.44 TULP2tubby like protein 2 (206733_at), score: 0.76 TWISTNBTWIST neighbor (214729_at), score: -0.42 TXNthioredoxin (216609_at), score: 0.53 TYMPthymidine phosphorylase (217497_at), score: 0.47 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: -0.42 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.6 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: 0.4 UPF1UPF1 regulator of nonsense transcripts homolog (yeast) (211168_s_at), score: -0.49 USP12ubiquitin specific peptidase 12 (213327_s_at), score: 0.41 USP19ubiquitin specific peptidase 19 (214674_at), score: -0.4 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.38 VAMP8vesicle-associated membrane protein 8 (endobrevin) (202546_at), score: 0.41 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.59 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.51 VGFVGF nerve growth factor inducible (205586_x_at), score: 0.44 VHLvon Hippel-Lindau tumor suppressor (203844_at), score: 0.47 VPS11vacuolar protein sorting 11 homolog (S. cerevisiae) (203292_s_at), score: 0.37 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.43 WDR52WD repeat domain 52 (221103_s_at), score: -0.39 WHSC1L1Wolf-Hirschhorn syndrome candidate 1-like 1 (218173_s_at), score: 0.42 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.37 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: 0.37 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (220020_at), score: -0.39 YDD19YDD19 protein (37079_at), score: -0.41 ZBTB24zinc finger and BTB domain containing 24 (205340_at), score: 0.46 ZBTB3zinc finger and BTB domain containing 3 (220391_at), score: -0.46 ZBTB33zinc finger and BTB domain containing 33 (214631_at), score: -0.48 ZC3H3zinc finger CCCH-type containing 3 (213445_at), score: -0.47 ZDHHC11zinc finger, DHHC-type containing 11 (221646_s_at), score: 0.46 ZFHX3zinc finger homeobox 3 (208033_s_at), score: -0.41 ZFYzinc finger protein, Y-linked (207247_s_at), score: -0.45 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: 0.44 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: 0.46 ZNF224zinc finger protein 224 (216983_s_at), score: 0.47 ZNF254zinc finger protein 254 (206862_at), score: -0.42 ZNF281zinc finger protein 281 (218401_s_at), score: 0.4 ZNF318zinc finger protein 318 (203521_s_at), score: -0.5 ZNF35zinc finger protein 35 (206096_at), score: 0.38 ZNF350zinc finger protein 350 (219266_at), score: 0.41 ZNF460zinc finger protein 460 (216279_at), score: -0.53 ZNF506zinc finger protein 506 (221626_at), score: 0.46 ZNF516zinc finger protein 516 (203604_at), score: 0.45 ZNF586zinc finger protein 586 (219711_at), score: 0.35 ZNF587zinc finger protein 587 (219981_x_at), score: 0.35 ZNF643zinc finger protein 643 (207219_at), score: -0.52 ZNF667zinc finger protein 667 (207120_at), score: 0.44 ZNF668zinc finger protein 668 (219047_s_at), score: -0.43 ZNF814zinc finger protein 814 (60794_f_at), score: -0.48 ZPBPzona pellucida binding protein (207021_at), score: 0.44 ZXDCZXD family zinc finger C (218639_s_at), score: 0.54
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |