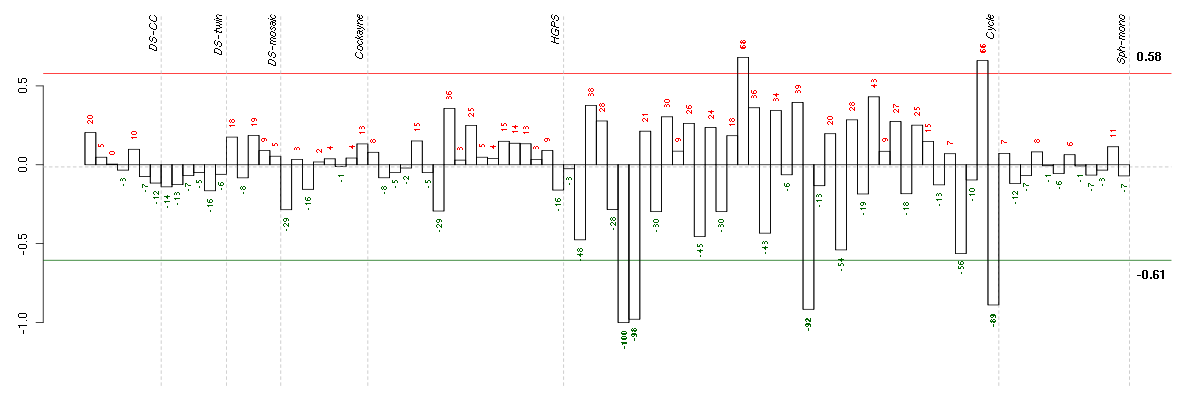

Under-expression is coded with green,
over-expression with red color.

signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.


ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.44 ABI3BPABI family, member 3 (NESH) binding protein (220518_at), score: 0.52 ABL1c-abl oncogene 1, receptor tyrosine kinase (202123_s_at), score: 0.53 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.62 ADAadenosine deaminase (204639_at), score: -0.51 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.65 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.95 AFG3L2AFG3 ATPase family gene 3-like 2 (yeast) (202486_at), score: 0.57 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.54 AGRNagrin (212285_s_at), score: 0.54 AK3L1adenylate kinase 3-like 1 (204348_s_at), score: 0.55 AK5adenylate kinase 5 (219308_s_at), score: -0.5 AKAP10A kinase (PRKA) anchor protein 10 (205045_at), score: -0.49 ANAPC10anaphase promoting complex subunit 10 (207845_s_at), score: -0.5 ANAPC2anaphase promoting complex subunit 2 (218555_at), score: 0.52 ANKRD28ankyrin repeat domain 28 (213035_at), score: -0.54 ANKRD36Bankyrin repeat domain 36B (220940_at), score: -0.5 ANO1anoctamin 1, calcium activated chloride channel (218804_at), score: 0.46 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (211975_at), score: 0.47 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: 0.48 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.67 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.57 ATF2activating transcription factor 2 (212984_at), score: -0.5 ATF5activating transcription factor 5 (204999_s_at), score: 0.55 ATF7IPactivating transcription factor 7 interacting protein (218987_at), score: -0.61 ATMataxia telangiectasia mutated (210858_x_at), score: -0.53 ATN1atrophin 1 (40489_at), score: 0.58 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.5 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: 0.71 ATXN1ataxin 1 (203232_s_at), score: -0.52 ATXN2ataxin 2 (202622_s_at), score: -0.54 AVPI1arginine vasopressin-induced 1 (218631_at), score: -0.55 AZI25-azacytidine induced 2 (218043_s_at), score: -0.53 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.64 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.5 BAHCC1BAH domain and coiled-coil containing 1 (219218_at), score: 0.49 BAIAP2BAI1-associated protein 2 (205294_at), score: 0.58 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.58 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.55 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (219528_s_at), score: 0.53 BCL2B-cell CLL/lymphoma 2 (203685_at), score: -0.52 BIN1bridging integrator 1 (210201_x_at), score: 0.68 BLCAPbladder cancer associated protein (201032_at), score: 0.46 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: 0.51 BRCA2breast cancer 2, early onset (208368_s_at), score: -0.65 BRD9bromodomain containing 9 (220155_s_at), score: 0.59 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.66 C10orf26chromosome 10 open reading frame 26 (202808_at), score: 0.53 C10orf76chromosome 10 open reading frame 76 (55662_at), score: -0.54 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.54 C12orf35chromosome 12 open reading frame 35 (218614_at), score: -0.51 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: -0.65 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.74 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.58 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: -0.48 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: 0.5 C1orf63chromosome 1 open reading frame 63 (209007_s_at), score: -0.53 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.58 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: 0.49 C22orf29chromosome 22 open reading frame 29 (219560_at), score: 0.48 C22orf9chromosome 22 open reading frame 9 (217118_s_at), score: 0.47 C4orf16chromosome 4 open reading frame 16 (219023_at), score: -0.5 C5orf28chromosome 5 open reading frame 28 (219029_at), score: -0.5 C7orf25chromosome 7 open reading frame 25 (53202_at), score: -0.5 C9orf9chromosome 9 open reading frame 9 (220050_at), score: 0.56 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.46 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.53 CASKIN2CASK interacting protein 2 (61297_at), score: 0.58 CCDC101coiled-coil domain containing 101 (48117_at), score: -0.48 CD248CD248 molecule, endosialin (219025_at), score: 0.5 CD70CD70 molecule (206508_at), score: -0.52 CDADC1cytidine and dCMP deaminase domain containing 1 (221015_s_at), score: -0.5 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.71 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.53 CDC42SE1CDC42 small effector 1 (218157_x_at), score: 0.48 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (209112_at), score: -0.54 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.71 CEP192centrosomal protein 192kDa (218827_s_at), score: -0.63 CEP57centrosomal protein 57kDa (203491_s_at), score: -0.61 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: 0.73 CICcapicua homolog (Drosophila) (212784_at), score: 0.58 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.55 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.51 CLCN2chloride channel 2 (213499_at), score: -0.61 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.51 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.51 CNN2calponin 2 (201605_x_at), score: 0.64 CNNM2cyclin M2 (206818_s_at), score: -0.49 CNNM3cyclin M3 (220739_s_at), score: 0.51 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.58 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.72 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.52 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: 0.61 CORINcorin, serine peptidase (220356_at), score: -0.66 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.55 CRKLv-crk sarcoma virus CT10 oncogene homolog (avian)-like (212180_at), score: 0.47 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.73 CSDE1cold shock domain containing E1, RNA-binding (202646_s_at), score: 0.46 CSPP1centrosome and spindle pole associated protein 1 (220072_at), score: -0.52 CTPS2CTP synthase II (219080_s_at), score: -0.51 CUX1cut-like homeobox 1 (214743_at), score: -0.55 CXCL11chemokine (C-X-C motif) ligand 11 (210163_at), score: 0.54 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.5 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.54 CYTH1cytohesin 1 (202880_s_at), score: 0.59 DBTdihydrolipoamide branched chain transacylase E2 (205370_x_at), score: -0.48 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: 0.58 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (219111_s_at), score: 0.47 DENND1ADENN/MADD domain containing 1A (219763_at), score: -0.51 DENND4BDENN/MADD domain containing 4B (202860_at), score: 0.68 DEXIdexamethasone-induced transcript (203733_at), score: 0.5 DIAPH2diaphanous homolog 2 (Drosophila) (205603_s_at), score: -0.58 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: -0.56 DLG3discs, large homolog 3 (Drosophila) (212728_at), score: 0.51 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: -0.53 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.57 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.81 DNA2DNA replication helicase 2 homolog (yeast) (213647_at), score: -0.48 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.55 DNM2dynamin 2 (202253_s_at), score: 0.44 DNM3dynamin 3 (209839_at), score: 0.55 DOK4docking protein 4 (209691_s_at), score: 0.68 DSC3desmocollin 3 (206032_at), score: 0.54 DTX4deltex homolog 4 (Drosophila) (212611_at), score: 0.5 DUSP6dual specificity phosphatase 6 (208891_at), score: 0.45 EDN1endothelin 1 (218995_s_at), score: 0.5 EEA1early endosome antigen 1 (204840_s_at), score: -0.57 EHHADHenoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase (205222_at), score: -0.65 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.54 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: 0.46 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: -0.56 EPHA3EPH receptor A3 (211164_at), score: 0.45 EPHA4EPH receptor A4 (206114_at), score: -0.55 EVLEnah/Vasp-like (217838_s_at), score: 0.47 F11coagulation factor XI (206610_s_at), score: -0.54 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.7 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.55 FAM3Afamily with sequence similarity 3, member A (38043_at), score: 0.51 FANCEFanconi anemia, complementation group E (220255_at), score: -0.49 FASNfatty acid synthase (212218_s_at), score: 0.65 FBLN2fibulin 2 (203886_s_at), score: 0.54 FBXL4F-box and leucine-rich repeat protein 4 (209943_at), score: -0.52 FBXO17F-box protein 17 (220233_at), score: 0.48 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.6 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.51 FKBPLFK506 binding protein like (219187_at), score: -0.6 FKRPfukutin related protein (219853_at), score: 0.72 FKSG2apoptosis inhibitor (208588_at), score: -0.58 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.87 FLJ20674hypothetical protein FLJ20674 (220137_at), score: -0.64 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.62 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.46 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: -0.51 FOXK2forkhead box K2 (203064_s_at), score: 0.94 FREQfrequenin homolog (Drosophila) (218266_s_at), score: 0.49 FRMPD4FERM and PDZ domain containing 4 (215052_at), score: -0.49 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.53 FZD5frizzled homolog 5 (Drosophila) (221245_s_at), score: -0.51 GALNAC4S-6STB cell RAG associated protein (203066_at), score: 0.48 GBLG protein beta subunit-like (220587_s_at), score: 0.48 GFPT2glutamine-fructose-6-phosphate transaminase 2 (205100_at), score: 0.49 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.5 GLI2GLI family zinc finger 2 (207034_s_at), score: 0.45 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.6 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.9 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: -0.65 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.65 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.5 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.62 HDAC5histone deacetylase 5 (202455_at), score: 0.5 HDAC7histone deacetylase 7 (217937_s_at), score: 0.54 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.49 HIST1H2BHhistone cluster 1, H2bh (208546_x_at), score: -0.48 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: -0.49 HLA-DMBmajor histocompatibility complex, class II, DM beta (203932_at), score: -0.49 HLXH2.0-like homeobox (214438_at), score: 0.55 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.49 HNRNPA3P1heterogeneous nuclear ribonucleoprotein A3 pseudogene 1 (206809_s_at), score: 0.52 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.5 HPS1Hermansky-Pudlak syndrome 1 (217354_s_at), score: 0.49 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.7 HSF1heat shock transcription factor 1 (202344_at), score: 0.5 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.54 ICAM1intercellular adhesion molecule 1 (202638_s_at), score: 0.52 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.64 IFRD1interferon-related developmental regulator 1 (202147_s_at), score: -0.55 IFT74intraflagellar transport 74 homolog (Chlamydomonas) (219174_at), score: -0.49 IL17RAinterleukin 17 receptor A (205707_at), score: 0.46 IL4Rinterleukin 4 receptor (203233_at), score: 0.53 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.47 INTS7integrator complex subunit 7 (222250_s_at), score: -0.5 IPO8importin 8 (205701_at), score: -0.48 IPPKinositol 1,3,4,5,6-pentakisphosphate 2-kinase (219092_s_at), score: 0.67 ITGA1integrin, alpha 1 (214660_at), score: -0.58 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: 0.5 JARID1Djumonji, AT rich interactive domain 1D (206700_s_at), score: -0.5 JMJD2Ajumonji domain containing 2A (203204_s_at), score: -0.58 JUNDjun D proto-oncogene (203751_x_at), score: 0.51 KAT2BK(lysine) acetyltransferase 2B (203845_at), score: -0.48 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: -0.65 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: 0.59 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.74 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.66 KIAA0317KIAA0317 (202128_at), score: 0.56 KIAA0556KIAA0556 (213185_at), score: 0.46 KIAA1009KIAA1009 (206005_s_at), score: -0.49 KIAA1305KIAA1305 (220911_s_at), score: 0.51 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.48 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.49 KRT24keratin 24 (220267_at), score: -0.5 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.5 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: 0.48 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.57 LOC100129250similar to hCG1811779 (78383_at), score: 0.5 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.46 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.57 LOC100133503similar to LRRC37A3 protein (220219_s_at), score: -0.51 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.49 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.52 LOC51152melanoma antigen (220771_at), score: 0.56 LOC643287similar to prothymosin alpha (216384_x_at), score: -0.49 LOC645139hypothetical LOC645139 (209064_x_at), score: 0.51 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: 0.45 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.56 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: 0.57 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.56 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.56 LRRC32leucine rich repeat containing 32 (203835_at), score: 0.45 LRRTM4leucine rich repeat transmembrane neuronal 4 (220345_at), score: 0.49 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: 0.57 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: 0.47 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.68 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.76 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: 0.45 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.53 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.55 MAPK8IP2mitogen-activated protein kinase 8 interacting protein 2 (205050_s_at), score: -0.55 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.64 MAPKAPK5mitogen-activated protein kinase-activated protein kinase 5 (212871_at), score: 0.45 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.58 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.6 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: -0.48 MC4Rmelanocortin 4 receptor (221467_at), score: -0.66 MCRS1microspherule protein 1 (202556_s_at), score: 0.48 MED15mediator complex subunit 15 (222175_s_at), score: 0.54 MED4mediator complex subunit 4 (217843_s_at), score: -0.53 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.56 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.62 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: 0.61 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.62 MKL1megakaryoblastic leukemia (translocation) 1 (212748_at), score: 0.48 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.61 MUC13mucin 13, cell surface associated (218687_s_at), score: -0.62 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.62 MYO9Bmyosin IXB (217297_s_at), score: 0.51 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.51 NCDNneurochondrin (209556_at), score: 0.52 NCKAP1NCK-associated protein 1 (207738_s_at), score: -0.54 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.46 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.48 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.46 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.58 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: 0.46 NEFMneurofilament, medium polypeptide (205113_at), score: -0.61 NELFnasal embryonic LHRH factor (221214_s_at), score: 0.46 NENFneuron derived neurotrophic factor (218407_x_at), score: 0.48 NF1neurofibromin 1 (211094_s_at), score: 0.46 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.61 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.55 NPATnuclear protein, ataxia-telangiectasia locus (209798_at), score: -0.52 NPC2Niemann-Pick disease, type C2 (200701_at), score: 0.52 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.5 NTRK3neurotrophic tyrosine kinase, receptor, type 3 (217033_x_at), score: 0.5 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: 0.81 NUPL2nucleoporin like 2 (204003_s_at), score: -0.66 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.56 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.67 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: 0.54 PAFAH1B2platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa (210160_at), score: -0.57 PARVBparvin, beta (37965_at), score: 0.57 PCDH11Yprotocadherin 11 Y-linked (217049_x_at), score: 0.45 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.5 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.64 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.5 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.57 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.5 PDE7Bphosphodiesterase 7B (220343_at), score: -0.49 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.48 PDK1pyruvate dehydrogenase kinase, isozyme 1 (206686_at), score: -0.48 PDLIM1PDZ and LIM domain 1 (208690_s_at), score: 0.45 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: 0.59 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.72 PHF2PHD finger protein 2 (212726_at), score: 0.44 PHF20PHD finger protein 20 (209422_at), score: -0.61 PHF7PHD finger protein 7 (215622_x_at), score: -0.5 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.59 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.64 PISDphosphatidylserine decarboxylase (202392_s_at), score: 0.59 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.46 PKN1protein kinase N1 (202161_at), score: 0.52 PKNOX1PBX/knotted 1 homeobox 1 (221883_at), score: -0.53 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.48 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: 0.62 PLCB4phospholipase C, beta 4 (203896_s_at), score: -0.49 PLCD1phospholipase C, delta 1 (205125_at), score: 0.62 PLCE1phospholipase C, epsilon 1 (205112_at), score: -0.5 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.6 PLEKHM2pleckstrin homology domain containing, family M (with RUN domain) member 2 (212146_at), score: 0.51 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.62 PMS2PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (209805_at), score: -0.69 POLHpolymerase (DNA directed), eta (219380_x_at), score: -0.54 POLIpolymerase (DNA directed) iota (219317_at), score: -0.51 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.49 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.66 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.79 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.52 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.52 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.47 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: 0.47 PRKCDprotein kinase C, delta (202545_at), score: 0.57 PRLRprolactin receptor (216638_s_at), score: 0.59 PRR5proline rich 5 (renal) (47069_at), score: 0.51 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.46 PTMAP7prothymosin, alpha pseudogene 7 (208549_x_at), score: -0.53 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.65 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.5 PXNpaxillin (211823_s_at), score: 0.57 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.63 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.47 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.77 RAB36RAB36, member RAS oncogene family (211471_s_at), score: 0.53 RAD17RAD17 homolog (S. pombe) (207405_s_at), score: -0.54 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.81 RALGDSral guanine nucleotide dissociation stimulator (209050_s_at), score: -0.48 RAP1BRAP1B, member of RAS oncogene family (200833_s_at), score: 0.46 RARAretinoic acid receptor, alpha (203749_s_at), score: 0.51 RBBP6retinoblastoma binding protein 6 (212781_at), score: -0.52 RBM15BRNA binding motif protein 15B (202689_at), score: 0.57 RCAN1regulator of calcineurin 1 (215253_s_at), score: -0.53 RDXradixin (204969_s_at), score: -0.51 REEP1receptor accessory protein 1 (204364_s_at), score: -0.48 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.57 RELBv-rel reticuloendotheliosis viral oncogene homolog B (205205_at), score: 0.47 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.68 RGS14regulator of G-protein signaling 14 (38290_at), score: 0.53 RGS2regulator of G-protein signaling 2, 24kDa (202388_at), score: 0.47 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.54 RNF187ring finger protein 187 (212155_at), score: 0.53 RNF19Bring finger protein 19B (213038_at), score: 0.44 RNF220ring finger protein 220 (219988_s_at), score: 0.57 RPL10P16ribosomal protein L10 pseudogene 16 (217680_x_at), score: -0.53 RPL15P22ribosomal protein L15 pseudogene 22 (217266_at), score: -0.49 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.7 RPL26P37ribosomal protein L26 pseudogene 37 (222229_x_at), score: -0.47 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.46 RPS11ribosomal protein S11 (213350_at), score: 0.51 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.71 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.48 S100A14S100 calcium binding protein A14 (218677_at), score: -0.5 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.73 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: 0.58 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: -0.52 SDC3syndecan 3 (202898_at), score: 0.52 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: 0.5 SF1splicing factor 1 (208313_s_at), score: 0.67 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.62 SH2B3SH2B adaptor protein 3 (203320_at), score: 0.56 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.55 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.49 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.58 SHQ1SHQ1 homolog (S. cerevisiae) (63009_at), score: 0.52 SHROOM2shroom family member 2 (204967_at), score: 0.59 SIM1single-minded homolog 1 (Drosophila) (206876_at), score: -0.53 SIP1survival of motor neuron protein interacting protein 1 (205063_at), score: -0.5 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.72 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.53 SLC25A22solute carrier family 25 (mitochondrial carrier: glutamate), member 22 (218725_at), score: 0.45 SLC6A8solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (202219_at), score: 0.55 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (207053_at), score: -0.48 SMARCD1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 (209518_at), score: 0.45 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (221516_s_at), score: 0.49 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.53 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.65 SMOXspermine oxidase (210357_s_at), score: 0.47 SMYD5SMYD family member 5 (209516_at), score: 0.56 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: -0.49 SNIP1Smad nuclear interacting protein 1 (219409_at), score: 0.48 SNX27sorting nexin family member 27 (221006_s_at), score: 0.62 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.91 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.55 SPATA2spermatogenesis associated 2 (204433_s_at), score: 0.48 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.59 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.48 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.57 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.53 ST7suppression of tumorigenicity 7 (207871_s_at), score: -0.62 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.51 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (207320_x_at), score: 0.56 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: -0.59 STK3serine/threonine kinase 3 (STE20 homolog, yeast) (204068_at), score: -0.56 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.53 SUPT5Hsuppressor of Ty 5 homolog (S. cerevisiae) (201480_s_at), score: 0.44 SUSD5sushi domain containing 5 (214954_at), score: -0.48 SYNPOsynaptopodin (202796_at), score: 0.45 SYTL2synaptotagmin-like 2 (220613_s_at), score: 0.48 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.72 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: 0.51 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.52 TBX2T-box 2 (40560_at), score: 0.63 TBX3T-box 3 (219682_s_at), score: 0.67 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.5 THADAthyroid adenoma associated (54632_at), score: -0.52 THUMPD1THUMP domain containing 1 (206555_s_at), score: -0.49 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.54 TLE2transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) (40837_at), score: 0.66 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.56 TLR3toll-like receptor 3 (206271_at), score: 0.46 TMCC1transmembrane and coiled-coil domain family 1 (213351_s_at), score: -0.5 TMEM123transmembrane protein 123 (211967_at), score: -0.51 TMEM149transmembrane protein 149 (219690_at), score: -0.6 TNFAIP3tumor necrosis factor, alpha-induced protein 3 (202644_s_at), score: 0.47 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.44 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.85 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: 0.52 TRIM21tripartite motif-containing 21 (204804_at), score: 0.45 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.52 TSKUtsukushin (218245_at), score: 1 TSPAN31tetraspanin 31 (203227_s_at), score: -0.49 TTBK2tau tubulin kinase 2 (213922_at), score: -0.53 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.53 TTTY15testis-specific transcript, Y-linked 15 (214983_at), score: 0.45 TWISTNBTWIST neighbor (214729_at), score: 0.52 TXLNAtaxilin alpha (212300_at), score: 0.69 UBAP1ubiquitin associated protein 1 (221490_at), score: 0.54 UBE2Zubiquitin-conjugating enzyme E2Z (217750_s_at), score: 0.49 UBE3Aubiquitin protein ligase E3A (213291_s_at), score: 0.47 UBR2ubiquitin protein ligase E3 component n-recognin 2 (212760_at), score: -0.48 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.67 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.48 USF2upstream transcription factor 2, c-fos interacting (214879_x_at), score: 0.53 USP36ubiquitin specific peptidase 36 (220370_s_at), score: 0.47 USP6NLUSP6 N-terminal like (204761_at), score: -0.55 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.66 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.74 WBP2WW domain binding protein 2 (209117_at), score: 0.48 WDR42AWD repeat domain 42A (202249_s_at), score: 0.69 WDR6WD repeat domain 6 (217734_s_at), score: 0.72 WIZwidely interspaced zinc finger motifs (52005_at), score: 0.6 WSB1WD repeat and SOCS box-containing 1 (201294_s_at), score: -0.5 WWTR1WW domain containing transcription regulator 1 (202132_at), score: -0.53 XAB2XPA binding protein 2 (218110_at), score: 0.59 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.6 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.67 ZBTB3zinc finger and BTB domain containing 3 (220391_at), score: -0.48 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: -0.72 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.47 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.72 ZMYM1zinc finger, MYM-type 1 (220206_at), score: -0.53 ZMYM5zinc finger, MYM-type 5 (206652_at), score: -0.48 ZNF212zinc finger protein 212 (203985_at), score: 0.47 ZNF248zinc finger protein 248 (213269_at), score: -0.59 ZNF277zinc finger protein 277 (218645_at), score: -0.48 ZNF280Azinc finger protein 280A (216034_at), score: 0.45 ZNF281zinc finger protein 281 (218401_s_at), score: -0.49 ZNF33Bzinc finger protein 33B (215022_x_at), score: -0.52 ZNF365zinc finger protein 365 (206448_at), score: 0.51 ZNF480zinc finger protein 480 (222283_at), score: -0.71 ZNF536zinc finger protein 536 (206403_at), score: 0.5 ZNF573zinc finger protein 573 (217627_at), score: -0.6 ZNF580zinc finger protein 580 (220748_s_at), score: 0.7 ZNF592zinc finger protein 592 (204473_s_at), score: 0.49 ZNF814zinc finger protein 814 (60794_f_at), score: 0.59 ZNF83zinc finger protein 83 (221645_s_at), score: -0.48
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |