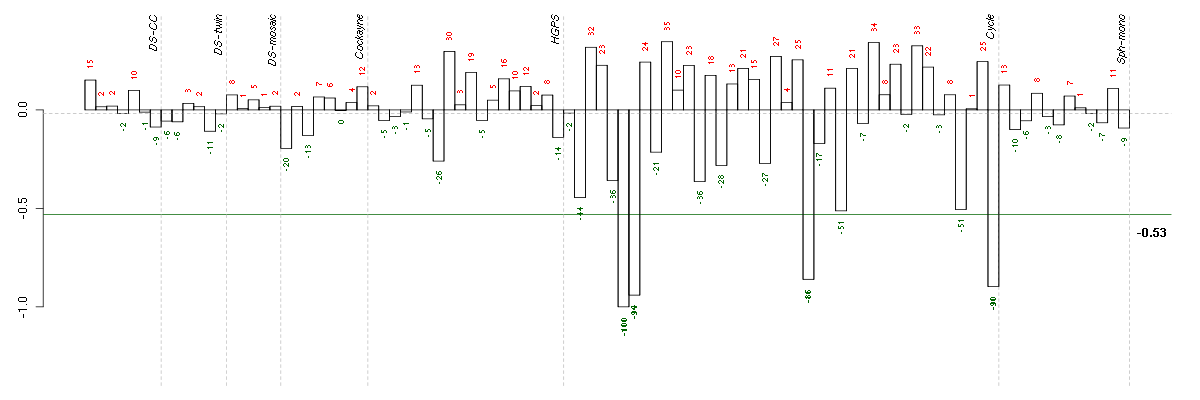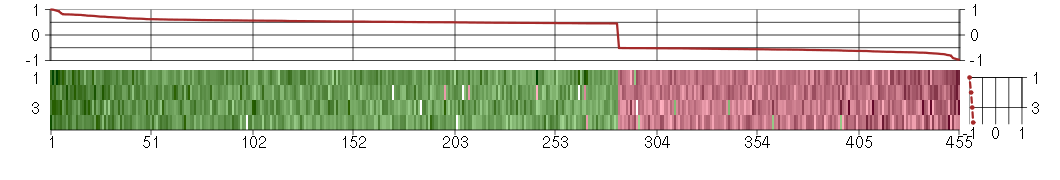

Under-expression is coded with green,
over-expression with red color.



ABHD2abhydrolase domain containing 2 (221815_at), score: -0.52 ABHD3abhydrolase domain containing 3 (213017_at), score: -0.55 ABI3BPABI family, member 3 (NESH) binding protein (220518_at), score: 0.5 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.56 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.55 ACTN3actinin, alpha 3 (206891_at), score: -0.56 ADAadenosine deaminase (204639_at), score: -0.6 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.58 ADCY3adenylate cyclase 3 (209321_s_at), score: 1 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.47 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.56 AGTPBP1ATP/GTP binding protein 1 (204500_s_at), score: -0.54 AK3L1adenylate kinase 3-like 1 (204348_s_at), score: 0.47 AK5adenylate kinase 5 (219308_s_at), score: -0.57 AKAP10A kinase (PRKA) anchor protein 10 (205045_at), score: -0.52 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: -0.57 ANAPC2anaphase promoting complex subunit 2 (218555_at), score: 0.6 ANGPTL2angiopoietin-like 2 (213004_at), score: 0.47 APLP1amyloid beta (A4) precursor-like protein 1 (209462_at), score: 0.49 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: 0.47 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (211975_at), score: 0.47 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: 0.58 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.52 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: 0.48 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.59 ATF7IPactivating transcription factor 7 interacting protein (218987_at), score: -0.52 ATMataxia telangiectasia mutated (210858_x_at), score: -0.56 ATN1atrophin 1 (40489_at), score: 0.58 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.52 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214149_s_at), score: 0.56 B3GNT1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (203188_at), score: -0.59 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: 0.48 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.51 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.53 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.47 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (219528_s_at), score: 0.53 BDH13-hydroxybutyrate dehydrogenase, type 1 (211715_s_at), score: -0.52 BIN1bridging integrator 1 (210201_x_at), score: 0.79 BLMBloom syndrome (205733_at), score: -0.54 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: 0.48 BRCA2breast cancer 2, early onset (208368_s_at), score: -0.79 BRD9bromodomain containing 9 (220155_s_at), score: 0.59 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: -0.53 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.66 BTG2BTG family, member 2 (201236_s_at), score: 0.53 C10orf26chromosome 10 open reading frame 26 (202808_at), score: 0.49 C10orf76chromosome 10 open reading frame 76 (55662_at), score: -0.52 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.57 C12orf35chromosome 12 open reading frame 35 (218614_at), score: -0.52 C13orf23chromosome 13 open reading frame 23 (218420_s_at), score: 0.49 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: -0.71 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.56 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.56 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: -0.62 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: 0.6 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.59 C20orf20chromosome 20 open reading frame 20 (218586_at), score: 0.48 C22orf9chromosome 22 open reading frame 9 (217118_s_at), score: 0.57 C4orf15chromosome 4 open reading frame 15 (210054_at), score: -0.63 C5orf28chromosome 5 open reading frame 28 (219029_at), score: -0.6 C7orf25chromosome 7 open reading frame 25 (53202_at), score: -0.62 C9orf127chromosome 9 open reading frame 127 (207839_s_at), score: 0.53 C9orf9chromosome 9 open reading frame 9 (220050_at), score: 0.61 CALB2calbindin 2 (205428_s_at), score: -0.61 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.6 CASP2caspase 2, apoptosis-related cysteine peptidase (209811_at), score: -0.55 CCDC101coiled-coil domain containing 101 (48117_at), score: -0.57 CD248CD248 molecule, endosialin (219025_at), score: 0.48 CD70CD70 molecule (206508_at), score: -0.64 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.67 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.46 CDC45LCDC45 cell division cycle 45-like (S. cerevisiae) (204126_s_at), score: -0.55 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.55 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.68 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: -0.51 CENPC1centromere protein C 1 (204739_at), score: -0.55 CENPJcentromere protein J (220885_s_at), score: -0.6 CENPQcentromere protein Q (219294_at), score: -0.68 CEP192centrosomal protein 192kDa (218827_s_at), score: -0.68 CEP57centrosomal protein 57kDa (203491_s_at), score: -0.67 CHKAcholine kinase alpha (204233_s_at), score: 0.46 CHPT1choline phosphotransferase 1 (221675_s_at), score: 0.46 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: 0.75 CICcapicua homolog (Drosophila) (212784_at), score: 0.6 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.54 CLCN2chloride channel 2 (213499_at), score: -0.56 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.54 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.54 CMTM6CKLF-like MARVEL transmembrane domain containing 6 (217947_at), score: 0.5 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.5 CNN2calponin 2 (201605_x_at), score: 0.46 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.62 CNR1cannabinoid receptor 1 (brain) (213436_at), score: 0.46 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.73 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.6 CORINcorin, serine peptidase (220356_at), score: -0.55 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.62 CPT1Bcarnitine palmitoyltransferase 1B (muscle) (210070_s_at), score: 0.53 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.71 CSN2casein beta (207951_at), score: 0.47 CSPP1centrosome and spindle pole associated protein 1 (220072_at), score: -0.52 CTPS2CTP synthase II (219080_s_at), score: -0.59 CUX1cut-like homeobox 1 (214743_at), score: -0.61 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.51 CXCL11chemokine (C-X-C motif) ligand 11 (210163_at), score: 0.5 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.56 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.53 DCTPP1dCTP pyrophosphatase 1 (218069_at), score: -0.58 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (212855_at), score: -0.53 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (219111_s_at), score: 0.46 DENND4BDENN/MADD domain containing 4B (202860_at), score: 0.81 DESdesmin (216947_at), score: 0.46 DEXIdexamethasone-induced transcript (203733_at), score: 0.54 DIAPH2diaphanous homolog 2 (Drosophila) (205603_s_at), score: -0.54 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: -0.67 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: -0.56 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.62 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.93 DNA2DNA replication helicase 2 homolog (yeast) (213647_at), score: -0.67 DNM1dynamin 1 (215116_s_at), score: 0.46 DNM3dynamin 3 (209839_at), score: 0.61 DOK4docking protein 4 (209691_s_at), score: 0.59 DSC3desmocollin 3 (206032_at), score: 0.6 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: 0.48 E2F8E2F transcription factor 8 (219990_at), score: -0.66 EEA1early endosome antigen 1 (204840_s_at), score: -0.58 EHHADHenoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase (205222_at), score: -0.51 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.57 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: 0.56 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: -0.68 EPHA4EPH receptor A4 (206114_at), score: -0.73 EXO1exonuclease 1 (204603_at), score: -0.52 F11coagulation factor XI (206610_s_at), score: -0.64 FAM119Bfamily with sequence similarity 119, member B (213861_s_at), score: 0.51 FAM131Afamily with sequence similarity 131, member A (221904_at), score: 0.49 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: 0.5 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.59 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.56 FAM3Afamily with sequence similarity 3, member A (38043_at), score: 0.56 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.48 FASNfatty acid synthase (212218_s_at), score: 0.73 FBLN2fibulin 2 (203886_s_at), score: 0.51 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: 0.55 FBXL4F-box and leucine-rich repeat protein 4 (209943_at), score: -0.55 FBXO17F-box protein 17 (220233_at), score: 0.6 FBXO2F-box protein 2 (219305_x_at), score: -0.52 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.54 FDXRferredoxin reductase (207813_s_at), score: 0.47 FKBPLFK506 binding protein like (219187_at), score: -0.74 FKRPfukutin related protein (219853_at), score: 0.69 FKSG2apoptosis inhibitor (208588_at), score: -0.65 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.81 FLJ20674hypothetical protein FLJ20674 (220137_at), score: -0.66 FMO4flavin containing monooxygenase 4 (206263_at), score: 0.51 FOXK2forkhead box K2 (203064_s_at), score: 0.97 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.52 GEMGTP binding protein overexpressed in skeletal muscle (204472_at), score: 0.46 GFPT2glutamine-fructose-6-phosphate transaminase 2 (205100_at), score: 0.52 GINS3GINS complex subunit 3 (Psf3 homolog) (45633_at), score: -0.62 GLRBglycine receptor, beta (205280_at), score: -0.55 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.69 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: 0.46 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.99 GORASP1golgi reassembly stacking protein 1, 65kDa (215749_s_at), score: 0.46 GP1BAglycoprotein Ib (platelet), alpha polypeptide (207389_at), score: -0.69 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.58 GPR1G protein-coupled receptor 1 (214605_x_at), score: -0.53 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: -0.57 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.47 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.57 H2BFSH2B histone family, member S (208579_x_at), score: -0.64 HDAC5histone deacetylase 5 (202455_at), score: 0.56 HDAC7histone deacetylase 7 (217937_s_at), score: 0.52 HECTD3HECT domain containing 3 (218632_at), score: 0.54 HERC5hect domain and RLD 5 (219863_at), score: 0.48 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.49 HIST1H2BEhistone cluster 1, H2be (208527_x_at), score: -0.62 HIST1H2BFhistone cluster 1, H2bf (208490_x_at), score: -0.58 HIST1H2BHhistone cluster 1, H2bh (208546_x_at), score: -0.72 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: -0.65 HNRNPA3P1heterogeneous nuclear ribonucleoprotein A3 pseudogene 1 (206809_s_at), score: 0.54 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.52 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.78 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.72 HTR2C5-hydroxytryptamine (serotonin) receptor 2C (211479_s_at), score: 0.47 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.59 IFI44interferon-induced protein 44 (214453_s_at), score: 0.49 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.57 INSIG1insulin induced gene 1 (201627_s_at), score: 0.58 INTS7integrator complex subunit 7 (222250_s_at), score: -0.55 ITGA1integrin, alpha 1 (214660_at), score: -0.63 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: 0.54 JMJD2Ajumonji domain containing 2A (203204_s_at), score: -0.53 KATNB1katanin p80 (WD repeat containing) subunit B 1 (203162_s_at), score: -0.72 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.47 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.75 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.61 KIAA0317KIAA0317 (202128_at), score: 0.59 KIAA0892KIAA0892 (212505_s_at), score: 0.51 KIAA1009KIAA1009 (206005_s_at), score: -0.66 KIAA1305KIAA1305 (220911_s_at), score: 0.69 KIAA1797KIAA1797 (218503_at), score: -0.52 KIF1Bkinesin family member 1B (209234_at), score: 0.48 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: 0.47 KLHDC10kelch domain containing 10 (209256_s_at), score: 0.5 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.54 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.53 KRT24keratin 24 (220267_at), score: -0.52 KRT7keratin 7 (209016_s_at), score: -0.59 LAMB4laminin, beta 4 (215516_at), score: 0.51 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: 0.57 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.72 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.71 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.56 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.75 LOC399491LOC399491 protein (214035_x_at), score: 0.58 LOC51152melanoma antigen (220771_at), score: 0.56 LOC643287similar to prothymosin alpha (216384_x_at), score: -0.54 LOC645139hypothetical LOC645139 (209064_x_at), score: 0.5 LOC65998hypothetical protein LOC65998 (218641_at), score: 0.46 LPCAT1lysophosphatidylcholine acyltransferase 1 (201818_at), score: 0.55 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.55 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: 0.62 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.52 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.65 LRRTM4leucine rich repeat transmembrane neuronal 4 (220345_at), score: 0.65 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: 0.53 LY75lymphocyte antigen 75 (205668_at), score: 0.48 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.46 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.69 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.49 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.51 MAPK13mitogen-activated protein kinase 13 (210058_at), score: -0.52 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.62 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.7 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: -0.58 MC4Rmelanocortin 4 receptor (221467_at), score: -0.93 MCRS1microspherule protein 1 (202556_s_at), score: 0.54 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: 0.47 MED15mediator complex subunit 15 (222175_s_at), score: 0.5 MED4mediator complex subunit 4 (217843_s_at), score: -0.59 MEF2Bmyocyte enhancer factor 2B (209926_at), score: -0.51 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.47 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.61 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: 0.57 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.59 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.52 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.65 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.52 MOAP1modulator of apoptosis 1 (212508_at), score: -0.52 MOCOSmolybdenum cofactor sulfurase (219959_at), score: -0.53 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.56 MR1major histocompatibility complex, class I-related (207565_s_at), score: -0.6 MRE11AMRE11 meiotic recombination 11 homolog A (S. cerevisiae) (205395_s_at), score: -0.52 MRPL2mitochondrial ribosomal protein L2 (218887_at), score: 0.57 MSL2male-specific lethal 2 homolog (Drosophila) (218733_at), score: 0.57 MSX2msh homeobox 2 (210319_x_at), score: 0.48 MTMR3myotubularin related protein 3 (202197_at), score: 0.53 MUC13mucin 13, cell surface associated (218687_s_at), score: -0.66 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.52 MYCL1v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian) (214058_at), score: 0.46 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.57 NAALAD2N-acetylated alpha-linked acidic dipeptidase 2 (222161_at), score: 0.54 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.5 NCDNneurochondrin (209556_at), score: 0.48 NCKAP1NCK-associated protein 1 (207738_s_at), score: -0.6 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.5 NEFMneurofilament, medium polypeptide (205113_at), score: -0.81 NENFneuron derived neurotrophic factor (218407_x_at), score: 0.57 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.64 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.52 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.55 NLGN1neuroligin 1 (205893_at), score: -0.6 NPATnuclear protein, ataxia-telangiectasia locus (209798_at), score: -0.57 NPC2Niemann-Pick disease, type C2 (200701_at), score: 0.46 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.7 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: 0.49 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: 0.74 NUPL2nucleoporin like 2 (204003_s_at), score: -0.63 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.48 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.55 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.49 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.59 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.68 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: 0.5 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.47 PAFAH1B2platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa (210160_at), score: -0.67 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: 0.48 PARVBparvin, beta (37965_at), score: 0.6 PCDH9protocadherin 9 (219737_s_at), score: -0.68 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.63 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.53 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.53 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.52 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.57 PDIA6protein disulfide isomerase family A, member 6 (208638_at), score: -0.52 PDK3pyruvate dehydrogenase kinase, isozyme 3 (221957_at), score: 0.5 PDLIM1PDZ and LIM domain 1 (208690_s_at), score: 0.54 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: 0.52 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.8 PHF20PHD finger protein 20 (209422_at), score: -0.54 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.77 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.58 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.53 PKN1protein kinase N1 (202161_at), score: 0.55 PKNOX1PBX/knotted 1 homeobox 1 (221883_at), score: -0.63 PLAGL2pleiomorphic adenoma gene-like 2 (202925_s_at), score: 0.55 PLCB4phospholipase C, beta 4 (203896_s_at), score: -0.51 PLCD1phospholipase C, delta 1 (205125_at), score: 0.6 PLCE1phospholipase C, epsilon 1 (205112_at), score: -0.58 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.47 PLEKHM2pleckstrin homology domain containing, family M (with RUN domain) member 2 (212146_at), score: 0.47 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.46 PMS2PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (209805_at), score: -0.66 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.48 POLHpolymerase (DNA directed), eta (219380_x_at), score: -0.62 POLQpolymerase (DNA directed), theta (219510_at), score: -0.55 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.47 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.52 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.78 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.53 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.5 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: 0.51 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: -0.55 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.63 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.53 PXNpaxillin (211823_s_at), score: 0.52 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.54 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.8 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.79 RAP1BRAP1B, member of RAS oncogene family (200833_s_at), score: 0.56 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: 0.47 RBBP9retinoblastoma binding protein 9 (221440_s_at), score: -0.55 RBM15BRNA binding motif protein 15B (202689_at), score: 0.52 RDXradixin (204969_s_at), score: -0.53 REEP1receptor accessory protein 1 (204364_s_at), score: -0.63 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.79 RNF187ring finger protein 187 (212155_at), score: 0.59 RNF220ring finger protein 220 (219988_s_at), score: 0.65 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.51 RPL10P16ribosomal protein L10 pseudogene 16 (217680_x_at), score: -0.59 RPL13Aribosomal protein L13a (200715_x_at), score: 0.46 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.75 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.5 RPS11ribosomal protein S11 (213350_at), score: 0.46 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.69 RRP12ribosomal RNA processing 12 homolog (S. cerevisiae) (216913_s_at), score: 0.47 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.6 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.46 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.65 SCAPERS phase cyclin A-associated protein in the ER (209741_x_at), score: -0.52 SCML1sex comb on midleg-like 1 (Drosophila) (218793_s_at), score: -0.63 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: 0.54 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: -0.58 SDC3syndecan 3 (202898_at), score: 0.48 SETBP1SET binding protein 1 (205933_at), score: 0.57 SF1splicing factor 1 (208313_s_at), score: 0.64 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.52 SGSM3small G protein signaling modulator 3 (214779_s_at), score: -0.56 SH2B3SH2B adaptor protein 3 (203320_at), score: 0.53 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.51 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.48 SHQ1SHQ1 homolog (S. cerevisiae) (63009_at), score: 0.53 SIPA1L3signal-induced proliferation-associated 1 like 3 (213600_at), score: 0.57 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.85 SLC19A1solute carrier family 19 (folate transporter), member 1 (211576_s_at), score: -0.53 SLC22A5solute carrier family 22 (organic cation/carnitine transporter), member 5 (205074_at), score: 0.49 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.57 SLC6A8solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (202219_at), score: 0.58 SMCHD1structural maintenance of chromosomes flexible hinge domain containing 1 (212569_at), score: -0.52 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (221516_s_at), score: 0.5 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.76 SMOXspermine oxidase (210357_s_at), score: 0.54 SMYD5SMYD family member 5 (209516_at), score: 0.5 SNX27sorting nexin family member 27 (221006_s_at), score: 0.79 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.81 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.49 SP2Sp2 transcription factor (204367_at), score: -0.6 SPATA2spermatogenesis associated 2 (204433_s_at), score: 0.53 SPATA20spermatogenesis associated 20 (218164_at), score: 0.46 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.58 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.57 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: 0.46 ST7suppression of tumorigenicity 7 (207871_s_at), score: -0.67 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.5 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (207320_x_at), score: 0.61 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: -0.69 STK3serine/threonine kinase 3 (STE20 homolog, yeast) (204068_at), score: -0.7 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.52 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.56 SUPT5Hsuppressor of Ty 5 homolog (S. cerevisiae) (201480_s_at), score: 0.46 SUSD5sushi domain containing 5 (214954_at), score: -0.54 SYNMsynemin, intermediate filament protein (212730_at), score: -0.55 SYTL2synaptotagmin-like 2 (220613_s_at), score: 0.51 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.67 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.56 TBX2T-box 2 (40560_at), score: 0.48 TBX3T-box 3 (219682_s_at), score: 0.76 TCEB3Btranscription elongation factor B polypeptide 3B (elongin A2) (220844_at), score: 0.48 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.56 THBDthrombomodulin (203887_s_at), score: 0.48 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.59 TLE2transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) (40837_at), score: 0.53 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: 0.46 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.7 TLR3toll-like receptor 3 (206271_at), score: 0.5 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.58 TMEM149transmembrane protein 149 (219690_at), score: -0.91 TMEM159transmembrane protein 159 (213272_s_at), score: -0.51 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.46 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: 0.56 TNFRSF25tumor necrosis factor receptor superfamily, member 25 (219423_x_at), score: 0.53 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.58 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.95 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.6 TREX1three prime repair exonuclease 1 (34689_at), score: -0.52 TRIM21tripartite motif-containing 21 (204804_at), score: 0.52 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.56 TSKUtsukushin (218245_at), score: 0.95 TSPAN31tetraspanin 31 (203227_s_at), score: -0.51 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.65 TTLL5tubulin tyrosine ligase-like family, member 5 (214672_at), score: -0.53 TTLL7tubulin tyrosine ligase-like family, member 7 (219882_at), score: -0.61 TXLNAtaxilin alpha (212300_at), score: 0.68 UBAP1ubiquitin associated protein 1 (221490_at), score: 0.57 UBP1upstream binding protein 1 (LBP-1a) (218082_s_at), score: 0.49 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.56 ULBP2UL16 binding protein 2 (221291_at), score: -0.61 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.5 VAV3vav 3 guanine nucleotide exchange factor (218807_at), score: 0.54 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.48 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.72 WDR26WD repeat domain 26 (218107_at), score: 0.5 WDR42AWD repeat domain 42A (202249_s_at), score: 0.56 WDR6WD repeat domain 6 (217734_s_at), score: 0.81 WIZwidely interspaced zinc finger motifs (52005_at), score: 0.5 WWTR1WW domain containing transcription regulator 1 (202132_at), score: -0.67 XAB2XPA binding protein 2 (218110_at), score: 0.62 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.6 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.76 ZAKsterile alpha motif and leucine zipper containing kinase AZK (218833_at), score: -0.63 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: -0.97 ZDHHC4zinc finger, DHHC-type containing 4 (220261_s_at), score: 0.46 ZFP106zinc finger protein 106 homolog (mouse) (217781_s_at), score: 0.58 ZMYM1zinc finger, MYM-type 1 (220206_at), score: -0.52 ZNF266zinc finger protein 266 (214686_at), score: -0.51 ZNF281zinc finger protein 281 (218401_s_at), score: -0.52 ZNF365zinc finger protein 365 (206448_at), score: 0.51 ZNF480zinc finger protein 480 (222283_at), score: -0.54 ZNF536zinc finger protein 536 (206403_at), score: 0.48 ZNF580zinc finger protein 580 (220748_s_at), score: 0.64 ZNF702Pzinc finger protein 702 pseudogene (206557_at), score: 0.52 ZNF814zinc finger protein 814 (60794_f_at), score: 0.56 ZNF83zinc finger protein 83 (221645_s_at), score: -0.56
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |