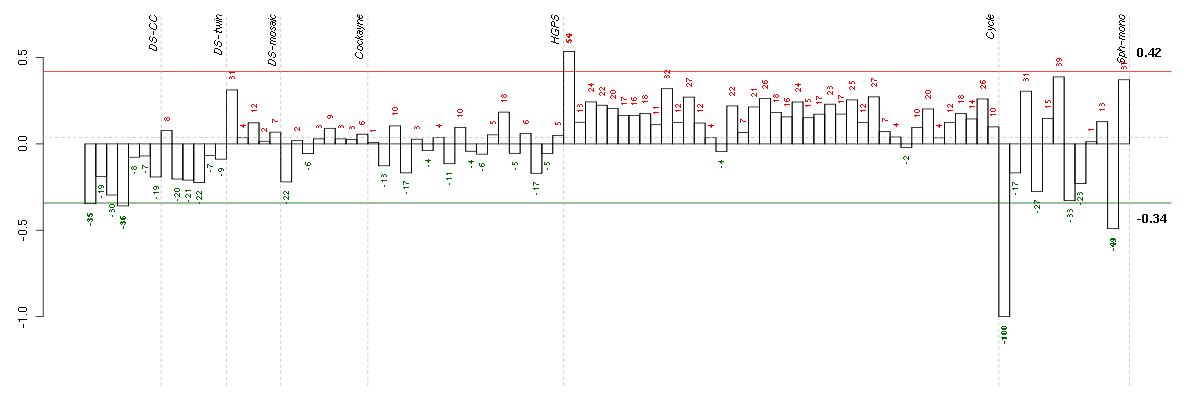

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 2.317e-03 | 3.337 | 13 | 78 | Neuroactive ligand-receptor interaction |
AASSaminoadipate-semialdehyde synthase (214829_at), score: -0.42 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (203504_s_at), score: -0.43 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: -0.7 ABI3BPABI family, member 3 (NESH) binding protein (220518_at), score: -0.35 ACOT11acyl-CoA thioesterase 11 (216103_at), score: -0.82 ACSL5acyl-CoA synthetase long-chain family member 5 (218322_s_at), score: -0.64 ACVR2Aactivin A receptor, type IIA (205327_s_at), score: -0.38 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: -0.51 ADAMTSL3ADAMTS-like 3 (213974_at), score: -0.55 ADAP2ArfGAP with dual PH domains 2 (219358_s_at), score: -0.44 ADCY2adenylate cyclase 2 (brain) (217687_at), score: -0.34 ADORA1adenosine A1 receptor (216220_s_at), score: -0.62 ADORA2Aadenosine A2a receptor (205013_s_at), score: -0.91 AIPL1aryl hydrocarbon receptor interacting protein-like 1 (219977_at), score: -0.35 AKIRIN2akirin 2 (213810_s_at), score: -0.38 ALDH1B1aldehyde dehydrogenase 1 family, member B1 (209646_x_at), score: -0.35 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: -0.74 ALMS1Alstrom syndrome 1 (214707_x_at), score: -0.95 ANKHankylosis, progressive homolog (mouse) (220076_at), score: -0.42 ANKRD10ankyrin repeat domain 10 (218093_s_at), score: -0.56 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: -0.78 APODapolipoprotein D (201525_at), score: -0.77 ARAP2ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 (214102_at), score: -0.65 ARG2arginase, type II (203945_at), score: -0.51 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: -0.51 ATP5OATP synthase, H+ transporting, mitochondrial F1 complex, O subunit (216954_x_at), score: -0.62 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: -0.56 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (221234_s_at), score: -0.69 BCL2L11BCL2-like 11 (apoptosis facilitator) (222343_at), score: -0.41 BCL9B-cell CLL/lymphoma 9 (204129_at), score: -0.81 BEANbrain expressed, associated with Nedd4 (214068_at), score: -0.4 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: -0.72 BTF3L2basic transcription factor 3, like 2 (217461_x_at), score: -0.43 C10orf110chromosome 10 open reading frame 110 (220703_at), score: -0.65 C11orf57chromosome 11 open reading frame 57 (218314_s_at), score: -0.42 C13orf23chromosome 13 open reading frame 23 (218420_s_at), score: -0.59 C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: -0.6 C17orf86chromosome 17 open reading frame 86 (221621_at), score: -0.67 C1orf135chromosome 1 open reading frame 135 (220011_at), score: -0.37 C20orf20chromosome 20 open reading frame 20 (218586_at), score: -0.57 C3complement component 3 (217767_at), score: -0.37 C4orf10chromosome 4 open reading frame 10 (214123_s_at), score: -0.46 C6orf106chromosome 6 open reading frame 106 (217925_s_at), score: -0.4 C6orf162chromosome 6 open reading frame 162 (213314_at), score: -0.42 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.55 CADM4cell adhesion molecule 4 (222293_at), score: -0.56 CADPS2Ca++-dependent secretion activator 2 (219572_at), score: -0.52 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.6 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: -0.58 CCR9chemokine (C-C motif) receptor 9 (207445_s_at), score: -0.47 CD28CD28 molecule (206545_at), score: -0.34 CD53CD53 molecule (203416_at), score: -0.47 CDC14ACDC14 cell division cycle 14 homolog A (S. cerevisiae) (205288_at), score: -0.72 CDKAL1CDK5 regulatory subunit associated protein 1-like 1 (214877_at), score: -0.45 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: -0.79 CDR1cerebellar degeneration-related protein 1, 34kDa (207276_at), score: -0.69 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: -0.63 CEP27centrosomal protein 27kDa (220071_x_at), score: -0.66 CGGBP1CGG triplet repeat binding protein 1 (214050_at), score: -1 CHD2chromodomain helicase DNA binding protein 2 (203461_at), score: -0.45 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: -0.77 CHN2chimerin (chimaerin) 2 (207486_x_at), score: -0.49 CIRBPcold inducible RNA binding protein (200811_at), score: -0.71 CISD3CDGSH iron sulfur domain 3 (213551_x_at), score: -0.55 CLEC4EC-type lectin domain family 4, member E (219859_at), score: -0.63 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.39 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: -0.37 CNOT6CCR4-NOT transcription complex, subunit 6 (217970_s_at), score: -0.45 CNOT8CCR4-NOT transcription complex, subunit 8 (202163_s_at), score: -0.34 COX5Bcytochrome c oxidase subunit Vb (213736_at), score: -0.49 CPEcarboxypeptidase E (201117_s_at), score: -0.66 CREBZFCREB/ATF bZIP transcription factor (202978_s_at), score: -0.36 CSF2RBcolony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) (205159_at), score: -0.36 CTSScathepsin S (202901_x_at), score: -0.48 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: -0.49 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.38 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: -0.46 CXCL2chemokine (C-X-C motif) ligand 2 (209774_x_at), score: -0.71 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: -0.42 CXorf21chromosome X open reading frame 21 (220252_x_at), score: -0.49 CXorf56chromosome X open reading frame 56 (219805_at), score: -0.37 CYBBcytochrome b-245, beta polypeptide (217431_x_at), score: -0.46 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (216809_at), score: -0.58 CYP11B1cytochrome P450, family 11, subfamily B, polypeptide 1 (214610_at), score: -0.4 CYP1A2cytochrome P450, family 1, subfamily A, polypeptide 2 (207608_x_at), score: -0.48 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: -0.66 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: -0.71 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.54 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: -1 DAZ1deleted in azoospermia 1 (216922_x_at), score: -0.74 DAZ3deleted in azoospermia 3 (208281_x_at), score: -0.67 DAZ4deleted in azoospermia 4 (216351_x_at), score: -0.77 DDX6DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (204909_at), score: -0.44 DESdesmin (216947_at), score: -0.52 DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (206752_s_at), score: -0.36 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: -0.62 DKFZp686O1327hypothetical gene supported by BC043549; BX648102 (216877_at), score: -0.88 DLG3discs, large homolog 3 (Drosophila) (212728_at), score: -0.36 DLX4distal-less homeobox 4 (208216_at), score: -0.56 DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: -0.77 DNAH7dynein, axonemal, heavy chain 7 (214222_at), score: -0.55 DNM3dynamin 3 (209839_at), score: -0.38 DTX4deltex homolog 4 (Drosophila) (212611_at), score: -0.45 DUSP8dual specificity phosphatase 8 (206374_at), score: -0.37 EEF1Deukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) (214395_x_at), score: -0.7 EGR1early growth response 1 (201694_s_at), score: -0.39 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: -0.37 EIF5A2eukaryotic translation initiation factor 5A2 (220198_s_at), score: -0.35 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: -0.81 ELL3elongation factor RNA polymerase II-like 3 (219517_at), score: -0.55 EPAGearly lymphoid activation protein (217050_at), score: -0.54 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: -0.7 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: -0.59 FAM106Afamily with sequence similarity 106, member A (220575_at), score: -0.55 FAM134Afamily with sequence similarity 134, member A (222129_at), score: -0.63 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: -0.63 FAM53Bfamily with sequence similarity 53, member B (203206_at), score: -0.48 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: -0.63 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: -0.62 FBXO41F-box protein 41 (44040_at), score: -0.69 FBXW12F-box and WD repeat domain containing 12 (215600_x_at), score: -0.67 FCARFc fragment of IgA, receptor for (211305_x_at), score: -0.6 FCER1AFc fragment of IgE, high affinity I, receptor for; alpha polypeptide (211734_s_at), score: -0.44 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: -0.37 FGD6FYVE, RhoGEF and PH domain containing 6 (219901_at), score: -0.83 FGFR2fibroblast growth factor receptor 2 (208229_at), score: -0.4 FGL1fibrinogen-like 1 (205305_at), score: -0.43 FKSG49FKSG49 (211454_x_at), score: -0.46 FLJ10038hypothetical protein FLJ10038 (205510_s_at), score: -0.54 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: -0.86 FLJ21369hypothetical protein FLJ21369 (220401_at), score: -0.59 FLJ23172hypothetical LOC389177 (217016_x_at), score: -0.74 FNBP4formin binding protein 4 (212232_at), score: -0.56 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: -0.57 FOXC1forkhead box C1 (213260_at), score: -0.5 FOXO1forkhead box O1 (202724_s_at), score: -0.37 FUBP1far upstream element (FUSE) binding protein 1 (212847_at), score: -0.54 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: -0.47 FUT3fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group) (214088_s_at), score: -0.64 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: -0.96 GCLCglutamate-cysteine ligase, catalytic subunit (202922_at), score: -0.48 GK2glycerol kinase 2 (215430_at), score: -0.62 GLRA1glycine receptor, alpha 1 (207972_at), score: -0.34 GNG4guanine nucleotide binding protein (G protein), gamma 4 (205184_at), score: -0.42 GNRHRgonadotropin-releasing hormone receptor (216341_s_at), score: -0.38 GOLGA8Agolgi autoantigen, golgin subfamily a, 8A (208798_x_at), score: -0.36 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: -0.38 GORASP1golgi reassembly stacking protein 1, 65kDa (56919_at), score: -0.4 GP6glycoprotein VI (platelet) (220336_s_at), score: -0.43 GPM6Bglycoprotein M6B (209167_at), score: -0.58 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: -0.59 GRM6glutamate receptor, metabotropic 6 (208035_at), score: -0.56 GTPBP3GTP binding protein 3 (mitochondrial) (213835_x_at), score: -0.55 GZMMgranzyme M (lymphocyte met-ase 1) (207460_at), score: -0.45 H2AFJH2A histone family, member J (220936_s_at), score: -0.53 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: -0.59 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.56 HBBhemoglobin, beta (211696_x_at), score: -0.42 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: -0.74 HECAheadcase homolog (Drosophila) (218603_at), score: -0.46 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.57 HIST2H2BEhistone cluster 2, H2be (202708_s_at), score: -0.5 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: -0.61 HMGCS13-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) (221750_at), score: -0.63 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (213359_at), score: -0.89 HTR1E5-hydroxytryptamine (serotonin) receptor 1E (207404_s_at), score: -0.37 HTR1F5-hydroxytryptamine (serotonin) receptor 1F (221458_at), score: -0.35 IBD12Inflammatory bowel disease 12 (215373_x_at), score: -0.9 IER3IP1immediate early response 3 interacting protein 1 (211406_at), score: -0.42 IFNA16interferon, alpha 16 (208448_x_at), score: -0.6 IFNA5interferon, alpha 5 (214569_at), score: -0.39 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: -0.53 IL1RNinterleukin 1 receptor antagonist (212657_s_at), score: -0.46 IMAASLC7A5 pseudogene (208118_x_at), score: -0.37 INSRinsulin receptor (213792_s_at), score: -0.79 ISL1ISL LIM homeobox 1 (206104_at), score: -0.56 ITGB5integrin, beta 5 (214021_x_at), score: -0.63 ITM2Aintegral membrane protein 2A (202746_at), score: -0.4 ITPKBinositol 1,4,5-trisphosphate 3-kinase B (203723_at), score: -0.58 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: -0.59 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: -0.35 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: -0.48 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: -0.52 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: -0.72 KERAkeratocan (220504_at), score: -0.35 KIAA0894KIAA0894 protein (207436_x_at), score: -0.51 KLF11Kruppel-like factor 11 (218486_at), score: -0.66 KLF9Kruppel-like factor 9 (203543_s_at), score: -0.35 LAMB4laminin, beta 4 (215516_at), score: -0.59 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: -0.4 LETM1leucine zipper-EF-hand containing transmembrane protein 1 (222006_at), score: -0.35 LHX3LIM homeobox 3 (221670_s_at), score: -0.34 LIMK1LIM domain kinase 1 (204357_s_at), score: -0.38 LOC100128701heterogeneous nuclear ribonucleoprotein A1-like 2 pseudogene (216497_at), score: -0.73 LOC100128836similar to heterogeneous nuclear ribonucleoprotein A1 (217353_at), score: -0.7 LOC100129624hypothetical LOC100129624 (210748_at), score: -0.72 LOC100130233similar to NPM1 protein (216387_x_at), score: -0.64 LOC100132214similar to HSPC047 protein (220692_at), score: -0.43 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: -0.51 LOC100134401hypothetical protein LOC100134401 (213605_s_at), score: -0.67 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.39 LOC152719hypothetical protein LOC152719 (215978_x_at), score: -0.4 LOC171220destrin-2 pseudogene (211325_x_at), score: -0.6 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: -0.85 LOC282997hypothetical protein LOC282997 (222307_at), score: -0.44 LOC440345hypothetical protein LOC440345 (214984_at), score: -0.34 LOC51152melanoma antigen (220771_at), score: -0.5 LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: -0.51 LOC644617hypothetical LOC644617 (221235_s_at), score: -0.51 LOC647070hypothetical LOC647070 (215467_x_at), score: -0.59 LOC728844hypothetical LOC728844 (222040_at), score: -0.78 LOC91316glucuronidase, beta/ immunoglobulin lambda-like polypeptide 1 pseudogene (215816_at), score: -0.53 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: -0.34 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: -0.58 LRRC37B2leucine rich repeat containing 37, member B2 (216149_at), score: -0.36 LSM14BLSM14B, SCD6 homolog B (S. cerevisiae) (219653_at), score: -0.35 MAGT1magnesium transporter 1 (210596_at), score: -0.39 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: -0.72 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: -0.77 MCL1myeloid cell leukemia sequence 1 (BCL2-related) (214057_at), score: -0.73 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: -0.36 MEFVMediterranean fever (208262_x_at), score: -0.76 MEP1Bmeprin A, beta (207251_at), score: -0.53 METAP2methionyl aminopeptidase 2 (202015_x_at), score: -0.87 METTL7Amethyltransferase like 7A (211424_x_at), score: -0.92 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.51 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: -0.64 MLLT10myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 (216506_x_at), score: -0.6 MMDmonocyte to macrophage differentiation-associated (203414_at), score: -0.35 MMRN2multimerin 2 (219091_s_at), score: -0.36 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.53 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: -0.47 MRPL44mitochondrial ribosomal protein L44 (218202_x_at), score: -0.7 MSL2male-specific lethal 2 homolog (Drosophila) (218733_at), score: -0.63 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.49 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.47 MXI1MAX interactor 1 (202364_at), score: -0.54 MYCL1v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian) (214058_at), score: -0.34 MYH15myosin, heavy chain 15 (215331_at), score: -0.37 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: -0.77 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: -0.42 NAT11N-acetyltransferase 11 (GCN5-related, putative) (222369_at), score: -0.38 NCRNA00092non-protein coding RNA 92 (215861_at), score: -0.58 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.51 NFYAnuclear transcription factor Y, alpha (204108_at), score: -0.53 NHLH2nescient helix loop helix 2 (215228_at), score: -0.52 NIPSNAP3Bnipsnap homolog 3B (C. elegans) (221104_s_at), score: -0.34 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: -0.63 NPY2Rneuropeptide Y receptor Y2 (210730_s_at), score: -0.39 NRCAMneuronal cell adhesion molecule (216959_x_at), score: -0.47 NUP214nucleoporin 214kDa (202155_s_at), score: -0.49 OLAHoleoyl-ACP hydrolase (219975_x_at), score: -0.39 OPHN1oligophrenin 1 (206323_x_at), score: -0.44 OR2B2olfactory receptor, family 2, subfamily B, member 2 (216408_at), score: -0.57 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.35 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.62 PARP8poly (ADP-ribose) polymerase family, member 8 (219033_at), score: -0.47 PATZ1POZ (BTB) and AT hook containing zinc finger 1 (209431_s_at), score: -0.48 PBX2pre-B-cell leukemia homeobox 2 (202876_s_at), score: -0.86 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: -0.48 PCGF2polycomb group ring finger 2 (203793_x_at), score: -0.48 PDE4Cphosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) (206792_x_at), score: -0.63 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.54 PEG3paternally expressed 3 (209242_at), score: -0.42 PELI2pellino homolog 2 (Drosophila) (219132_at), score: -0.73 PER2period homolog 2 (Drosophila) (205251_at), score: -0.59 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (202464_s_at), score: -0.41 PGFplacental growth factor (215179_x_at), score: -0.55 PHF21APHD finger protein 21A (203278_s_at), score: -0.47 PHF8PHD finger protein 8 (212916_at), score: -0.5 PHLPPPH domain and leucine rich repeat protein phosphatase (212719_at), score: -0.41 PIM2pim-2 oncogene (204269_at), score: -0.45 PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: -0.36 PIP5K1Bphosphatidylinositol-4-phosphate 5-kinase, type I, beta (217477_at), score: -0.39 PLA2G7phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) (206214_at), score: -0.41 PLCL2phospholipase C-like 2 (213309_at), score: -0.42 PLGLB1plasminogen-like B1 (214415_at), score: -0.4 PLS1plastin 1 (I isoform) (205190_at), score: -0.39 PMP2peripheral myelin protein 2 (206826_at), score: -0.42 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.36 PODNL1podocan-like 1 (220411_x_at), score: -0.37 POGZpogo transposable element with ZNF domain (215281_x_at), score: -0.9 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: -0.56 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: -0.65 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.37 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: -0.61 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: -0.38 PPP1R14Dprotein phosphatase 1, regulatory (inhibitor) subunit 14D (220082_at), score: -0.51 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: -0.98 PROL1proline rich, lacrimal 1 (208004_at), score: -0.4 PRPF39PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae) (220553_s_at), score: -0.35 PTGESprostaglandin E synthase (210367_s_at), score: -0.52 PTP4A3protein tyrosine phosphatase type IVA, member 3 (209695_at), score: -0.33 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: -0.49 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: -0.44 RAB11FIP1RAB11 family interacting protein 1 (class I) (219681_s_at), score: -0.51 RAB26RAB26, member RAS oncogene family (50965_at), score: -0.43 RAD52RAD52 homolog (S. cerevisiae) (205647_at), score: -0.4 RASL11BRAS-like, family 11, member B (219142_at), score: -0.42 RBMX2RNA binding motif protein, X-linked 2 (204098_at), score: -0.42 RETret proto-oncogene (205879_x_at), score: -0.33 RGL1ral guanine nucleotide dissociation stimulator-like 1 (209568_s_at), score: -0.37 RGS10regulator of G-protein signaling 10 (214000_s_at), score: -0.54 RGS5regulator of G-protein signaling 5 (218353_at), score: -0.78 RHEBRas homolog enriched in brain (213409_s_at), score: -0.55 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: -0.61 RLN1relaxin 1 (211753_s_at), score: -0.36 RNMTRNA (guanine-7-) methyltransferase (202683_s_at), score: -0.53 RP11-35N6.1plasticity related gene 3 (219732_at), score: -0.41 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.61 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.59 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.35 RPL14ribosomal protein L14 (219138_at), score: -0.53 RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: -0.81 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: -0.79 RPL35Aribosomal protein L35a (215208_x_at), score: -0.78 RPL37Aribosomal protein L37a (214041_x_at), score: -0.53 RPL38ribosomal protein L38 (202028_s_at), score: -0.34 RPLP2ribosomal protein, large, P2 (200908_s_at), score: -0.69 RPS10P3ribosomal protein S10 pseudogene 3 (217336_at), score: -0.7 RPS19ribosomal protein S19 (202648_at), score: -0.48 RPS3AP47ribosomal protein S3a pseudogene 47 (216823_at), score: -0.59 SAMD14sterile alpha motif domain containing 14 (213866_at), score: -0.34 SAP30LSAP30-like (219129_s_at), score: -0.49 SAPS2SAPS domain family, member 2 (202792_s_at), score: -0.43 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.46 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: -0.81 SCD5stearoyl-CoA desaturase 5 (220232_at), score: -0.57 SCML2sex comb on midleg-like 2 (Drosophila) (206147_x_at), score: -0.56 SCN11Asodium channel, voltage-gated, type XI, alpha subunit (220791_x_at), score: -0.6 SERF1Bsmall EDRK-rich factor 1B (centromeric) (219982_s_at), score: -0.79 SERPINA7serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (206386_at), score: -0.56 SERPINB13serpin peptidase inhibitor, clade B (ovalbumin), member 13 (211362_s_at), score: -0.48 SETD1ASET domain containing 1A (213202_at), score: -0.34 SFNstratifin (33322_i_at), score: -0.46 SFRS11splicing factor, arginine/serine-rich 11 (213742_at), score: -0.62 SFRS18splicing factor, arginine/serine-rich 18 (212176_at), score: -0.48 SFTPBsurfactant protein B (214354_x_at), score: -0.63 SH3BP2SH3-domain binding protein 2 (217257_at), score: -0.91 SH3GL3SH3-domain GRB2-like 3 (211565_at), score: -0.74 SHOX2short stature homeobox 2 (210135_s_at), score: -0.5 SIRPB1signal-regulatory protein beta 1 (206934_at), score: -0.51 SKP1S-phase kinase-associated protein 1 (200719_at), score: -0.51 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.45 SLC11A1solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 (217473_x_at), score: -0.63 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: -0.45 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: -0.44 SLC17A6solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (220551_at), score: -0.69 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: -0.86 SLC35F2solute carrier family 35, member F2 (218826_at), score: -0.54 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: -0.46 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (205921_s_at), score: -0.65 SMEK2SMEK homolog 2, suppressor of mek1 (Dictyostelium) (222270_at), score: -0.59 SNRPEsmall nuclear ribonucleoprotein polypeptide E (215450_at), score: -0.37 SNX15sorting nexin 15 (205482_x_at), score: -0.36 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: -0.42 SOX11SRY (sex determining region Y)-box 11 (204914_s_at), score: -0.35 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: -0.34 SPATA2spermatogenesis associated 2 (204433_s_at), score: -0.41 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: -0.46 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: -0.82 SPNsialophorin (206057_x_at), score: -0.82 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: -0.36 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: -0.53 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: -0.96 ST3GAL2ST3 beta-galactoside alpha-2,3-sialyltransferase 2 (217650_x_at), score: -0.48 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.38 STC1stanniocalcin 1 (204597_x_at), score: -0.39 SYCP1synaptonemal complex protein 1 (206740_x_at), score: -0.8 TAAR2trace amine associated receptor 2 (221394_at), score: -0.47 TAC1tachykinin, precursor 1 (206552_s_at), score: -0.49 TACR1tachykinin receptor 1 (208048_at), score: -0.38 TAS2R16taste receptor, type 2, member 16 (221444_at), score: -0.56 TATtyrosine aminotransferase (206916_x_at), score: -0.43 TBX6T-box 6 (207684_at), score: -0.57 TCTN2tectonic family member 2 (206438_x_at), score: -0.54 TFAP2Ctranscription factor AP-2 gamma (activating enhancer binding protein 2 gamma) (205286_at), score: -0.47 TGFB2transforming growth factor, beta 2 (220407_s_at), score: -0.51 TIGD1Ltigger transposable element derived 1-like (216459_x_at), score: -0.42 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: -0.85 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: -0.52 TMEM38Btransmembrane protein 38B (218772_x_at), score: -0.41 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: -0.45 TNFRSF9tumor necrosis factor receptor superfamily, member 9 (207536_s_at), score: -0.48 TNPO3transportin 3 (212317_at), score: -0.63 TP53TG3TP53 target 3 (220167_s_at), score: -0.82 TP63tumor protein p63 (209863_s_at), score: -0.41 TPTEtransmembrane phosphatase with tensin homology (220205_at), score: -0.58 TRA@T cell receptor alpha locus (216540_at), score: -0.49 TRA2Atransformer 2 alpha homolog (Drosophila) (204658_at), score: -0.38 TRIM36tripartite motif-containing 36 (219736_at), score: -0.98 TRPM3transient receptor potential cation channel, subfamily M, member 3 (220463_at), score: -0.34 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: -0.67 TSHBthyroid stimulating hormone, beta (214529_at), score: -0.36 TUBA3Ctubulin, alpha 3c (210527_x_at), score: -0.68 TULP4tubby like protein 4 (218184_at), score: -0.43 TXNthioredoxin (216609_at), score: -0.39 UBQLN4ubiquilin 4 (222252_x_at), score: -0.69 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: -0.83 UPP1uridine phosphorylase 1 (203234_at), score: -0.39 USP12ubiquitin specific peptidase 12 (213327_s_at), score: -0.39 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: -0.73 UTF1undifferentiated embryonic cell transcription factor 1 (208275_x_at), score: -0.49 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: -0.87 VEZTvezatin, adherens junctions transmembrane protein (207263_x_at), score: -0.66 VPS33Avacuolar protein sorting 33 homolog A (S. cerevisiae) (204590_x_at), score: -0.41 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: -0.58 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: -0.67 XYLT2xylosyltransferase II (219401_at), score: -0.4 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: -0.91 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: -0.82 ZEB2zinc finger E-box binding homeobox 2 (203603_s_at), score: -0.42 ZFC3H1zinc finger, C3H1-type containing (213065_at), score: -0.4 ZIC1Zic family member 1 (odd-paired homolog, Drosophila) (206373_at), score: -0.36 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: -0.46 ZNF117zinc finger protein 117 (207605_x_at), score: -0.51 ZNF160zinc finger protein 160 (214715_x_at), score: -0.59 ZNF432zinc finger protein 432 (219848_s_at), score: -0.42 ZNF440zinc finger protein 440 (215892_at), score: -0.65 ZNF451zinc finger protein 451 (212557_at), score: -0.34 ZNF492zinc finger protein 492 (215532_x_at), score: -0.83 ZNF493zinc finger protein 493 (211064_at), score: -0.65 ZNF506zinc finger protein 506 (221626_at), score: -0.45 ZNF528zinc finger protein 528 (215019_x_at), score: -0.51 ZNF552zinc finger protein 552 (219741_x_at), score: -0.51 ZNF611zinc finger protein 611 (208137_x_at), score: -0.39 ZNF639zinc finger protein 639 (218413_s_at), score: -0.69 ZNF701zinc finger protein 701 (220242_x_at), score: -0.53 ZNF747zinc finger protein 747 (206180_x_at), score: -0.55 ZNF770zinc finger protein 770 (220608_s_at), score: -0.66 ZNF816Azinc finger protein 816A (217541_x_at), score: -0.87 ZPBPzona pellucida binding protein (207021_at), score: -0.47 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: -0.53
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |