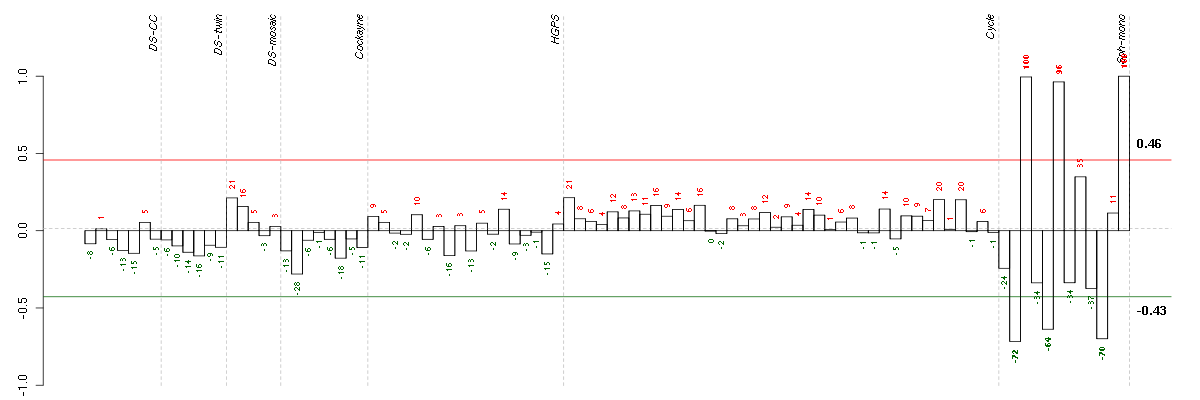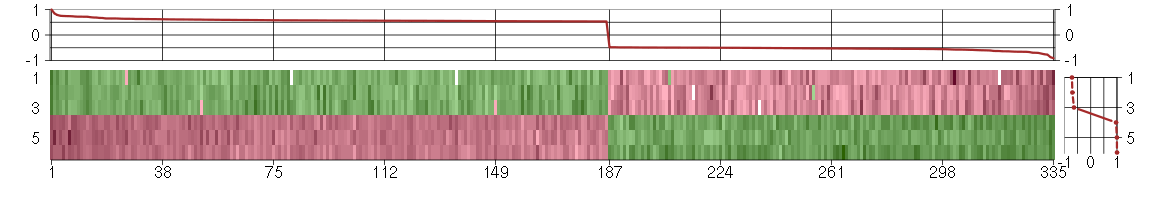

Under-expression is coded with green,
over-expression with red color.

eye development
The process whose specific outcome is the progression of the eye over time, from its formation to the mature structure. The eye is the organ of sight.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
negative regulation of immune system process
Any process that stops, prevents, or reduces the frequency, rate, or extent of an immune system process.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
embryonic development
The process whose specific outcome is the progression of an embryo from its formation until the end of its embryonic life stage. The end of the embryonic stage is organism-specific. For example, for mammals, the process would begin with zygote formation and end with birth. For insects, the process would begin at zygote formation and end with larval hatching. For plant zygotic embryos, this would be from zygote formation to the end of seed dormancy. For plant vegetative embryos, this would be from the initial determination of the cell or group of cells to form an embryo until the point when the embryo becomes independent of the parent plant.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
sensory organ development
The process whose specific outcome is the progression of sensory organs over time, from its formation to the mature structure.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
embryonic eye morphogenesis
The process occurring in the embryo by which the anatomical structures of the post-embryonic eye are generated and organized. Morphogenesis pertains to the creation of form.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ development
Development, taking place during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
negative regulation of response to stimulus
Any process that stops, prevents or reduces the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
eye morphogenesis
The process by which the anatomical structures of the eye are generated and organized. Morphogenesis pertains to the creation of form.
embryonic morphogenesis
The process by which anatomical structures are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form. The embryonic phase begins with zygote formation. The end of the embryonic phase is organism-specific. For example, it would be at birth for mammals, larval hatching for insects and seed dormancy in plants.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
negative regulation of immune response
Any process that stops, prevents or reduces the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
negative regulation of immune system process
Any process that stops, prevents, or reduces the frequency, rate, or extent of an immune system process.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
negative regulation of response to stimulus
Any process that stops, prevents or reduces the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of immune system process
Any process that stops, prevents, or reduces the frequency, rate, or extent of an immune system process.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
negative regulation of immune response
Any process that stops, prevents or reduces the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
embryonic development
The process whose specific outcome is the progression of an embryo from its formation until the end of its embryonic life stage. The end of the embryonic stage is organism-specific. For example, for mammals, the process would begin with zygote formation and end with birth. For insects, the process would begin at zygote formation and end with larval hatching. For plant zygotic embryos, this would be from zygote formation to the end of seed dormancy. For plant vegetative embryos, this would be from the initial determination of the cell or group of cells to form an embryo until the point when the embryo becomes independent of the parent plant.
embryonic morphogenesis
The process by which anatomical structures are generated and organized during the embryonic phase. Morphogenesis pertains to the creation of form. The embryonic phase begins with zygote formation. The end of the embryonic phase is organism-specific. For example, it would be at birth for mammals, larval hatching for insects and seed dormancy in plants.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
negative regulation of immune response
Any process that stops, prevents or reduces the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
negative regulation of response to stimulus
Any process that stops, prevents or reduces the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
negative regulation of immune response
Any process that stops, prevents or reduces the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ morphogenesis
Morphogenesis, during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic organ development
Development, taking place during the embryonic phase, of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
embryonic eye morphogenesis
The process occurring in the embryo by which the anatomical structures of the post-embryonic eye are generated and organized. Morphogenesis pertains to the creation of form.
eye morphogenesis
The process by which the anatomical structures of the eye are generated and organized. Morphogenesis pertains to the creation of form.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
steroid dehydrogenase activity
Catalysis of an oxidation-reduction (redox) reaction in which one substrate is a sterol derivative.
oxidoreductase activity, acting on CH-OH group of donors
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group act as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group acts as a hydrogen or electron donor and reduces NAD+ or NADP.
oxidoreductase activity, acting on the CH-CH group of donors
Catalysis of an oxidation-reduction (redox) reaction in which a CH-CH group acts as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which a CH-CH group acts as a hydrogen or electron donor and reduces NAD or NADP.
steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group acts as a hydrogen or electron donor and reduces NAD+ or NADP, and in which one substrate is a sterol derivative.
trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity
Catalysis of the reaction: NADP+ + trans-1,2-dihydrobenzene-1,2-diol = NADPH + catechol.
all
This term is the most general term possible
steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which a CH-OH group acts as a hydrogen or electron donor and reduces NAD+ or NADP, and in which one substrate is a sterol derivative.
ABAT4-aminobutyrate aminotransferase (209459_s_at), score: 0.54 ABHD2abhydrolase domain containing 2 (221815_at), score: -0.49 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.59 ACOT11acyl-CoA thioesterase 11 (216103_at), score: -0.59 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: 0.57 AEBP1AE binding protein 1 (201792_at), score: 0.54 AHI1Abelson helper integration site 1 (221569_at), score: 0.55 AIDAaxin interactor, dorsalization associated (220199_s_at), score: 0.56 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: 0.56 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.73 AKR1C1aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) (204151_x_at), score: -0.53 AKR1C2aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (209699_x_at), score: -0.54 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.52 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: 0.68 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: -0.5 ALDH7A1aldehyde dehydrogenase 7 family, member A1 (208951_at), score: 0.56 ANKHD1ankyrin repeat and KH domain containing 1 (208773_s_at), score: -0.5 ANXA8L2annexin A8-like 2 (203074_at), score: -0.53 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: -0.51 ARF4ADP-ribosylation factor 4 (201096_s_at), score: 0.54 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: 0.6 ARL15ADP-ribosylation factor-like 15 (219842_at), score: 0.64 ARL6IP5ADP-ribosylation-like factor 6 interacting protein 5 (200760_s_at), score: 0.53 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: 0.61 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: -0.49 ASPNasporin (219087_at), score: 0.6 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.53 ATP1A1ATPase, Na+/K+ transporting, alpha 1 polypeptide (220948_s_at), score: -0.5 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (212135_s_at), score: 0.53 BAGEB melanoma antigen (207712_at), score: -0.49 BEX1brain expressed, X-linked 1 (218332_at), score: 0.54 BGNbiglycan (213905_x_at), score: 0.59 BIRC2baculoviral IAP repeat-containing 2 (202076_at), score: -0.52 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: -0.6 BMP7bone morphogenetic protein 7 (209590_at), score: -0.53 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.59 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.56 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.72 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: 0.56 C20orf39chromosome 20 open reading frame 39 (219310_at), score: -0.58 C6orf120chromosome 6 open reading frame 120 (221786_at), score: 0.54 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.57 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: 0.54 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: 0.55 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.72 CD24CD24 molecule (266_s_at), score: -0.66 CD248CD248 molecule, endosialin (219025_at), score: 0.58 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.58 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: -0.69 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.7 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: 0.57 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.51 COBLL1COBL-like 1 (203642_s_at), score: 0.56 COL11A1collagen, type XI, alpha 1 (37892_at), score: 0.55 COL3A1collagen, type III, alpha 1 (215076_s_at), score: 0.63 CORO2Acoronin, actin binding protein, 2A (205538_at), score: -0.54 CTSScathepsin S (202901_x_at), score: -0.49 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.56 CYB561cytochrome b-561 (209163_at), score: -0.5 CYP2B6cytochrome P450, family 2, subfamily B, polypeptide 6 (217133_x_at), score: -0.55 D4S234EDNA segment on chromosome 4 (unique) 234 expressed sequence (209569_x_at), score: -0.5 DAPK1death-associated protein kinase 1 (203139_at), score: 0.6 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: -0.49 DAZ4deleted in azoospermia 4 (216351_x_at), score: -0.49 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.67 DKFZp547G183hypothetical LOC55525 (220572_at), score: -0.52 DLX4distal-less homeobox 4 (208216_at), score: -0.61 DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: -0.57 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: 0.6 DPY19L4dpy-19-like 4 (C. elegans) (213391_at), score: 0.54 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.61 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: -0.51 DSEdermatan sulfate epimerase (218854_at), score: 0.58 EIF3Deukaryotic translation initiation factor 3, subunit D (200005_at), score: -0.55 EIF4A1eukaryotic translation initiation factor 4A, isoform 1 (211787_s_at), score: -0.53 ELF1E74-like factor 1 (ets domain transcription factor) (212418_at), score: 0.59 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.53 ENOSF1enolase superfamily member 1 (204142_at), score: 0.56 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (209392_at), score: 0.62 EPAS1endothelial PAS domain protein 1 (200878_at), score: 0.57 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: -0.51 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: -0.52 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.56 ERGIC3ERGIC and golgi 3 (216032_s_at), score: -0.49 EVI2Aecotropic viral integration site 2A (204774_at), score: -0.54 EXOC7exocyst complex component 7 (212034_s_at), score: 0.53 FAF1Fas (TNFRSF6) associated factor 1 (218080_x_at), score: 0.57 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.52 FAM131Afamily with sequence similarity 131, member A (221904_at), score: -0.5 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.84 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: 0.74 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.72 FCARFc fragment of IgA, receptor for (211305_x_at), score: -0.51 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: -0.55 FCHSD2FCH and double SH3 domains 2 (203620_s_at), score: 0.56 FGF13fibroblast growth factor 13 (205110_s_at), score: -0.55 FKSG49FKSG49 (211454_x_at), score: -0.54 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: -0.55 FLJ42627hypothetical LOC645644 (220352_x_at), score: -0.54 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: 0.65 FOSL2FOS-like antigen 2 (218880_at), score: 0.54 FOXG1forkhead box G1 (206018_at), score: -0.53 FOXL2forkhead box L2 (220102_at), score: -0.51 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.58 FZD6frizzled homolog 6 (Drosophila) (203987_at), score: 0.6 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: -0.56 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: -0.48 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.67 GALCgalactosylceramidase (204417_at), score: 0.63 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (206335_at), score: -0.49 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.64 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.6 GIPC2GIPC PDZ domain containing family, member 2 (219970_at), score: 0.59 GLUD1glutamate dehydrogenase 1 (200946_x_at), score: 0.56 GMFBglia maturation factor, beta (202543_s_at), score: 0.56 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: 0.56 GON4Lgon-4-like (C. elegans) (218873_at), score: -0.49 GPR126G protein-coupled receptor 126 (213094_at), score: 0.65 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.52 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.6 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 1 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: -0.5 HEXBhexosaminidase B (beta polypeptide) (201944_at), score: -0.5 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.65 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.57 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: -0.5 HSPA2heat shock 70kDa protein 2 (211538_s_at), score: 0.54 HTATIP2HIV-1 Tat interactive protein 2, 30kDa (209448_at), score: 0.56 HYPKHuntingtin interacting protein K (218680_x_at), score: -0.54 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.76 IL10RBinterleukin 10 receptor, beta (209575_at), score: -0.49 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.72 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.55 ISL1ISL LIM homeobox 1 (206104_at), score: 0.62 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (213416_at), score: 0.62 ITPKBinositol 1,4,5-trisphosphate 3-kinase B (203723_at), score: 0.56 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (206765_at), score: 0.67 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.62 KIAA1324KIAA1324 (221874_at), score: -0.5 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.51 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.87 L1CAML1 cell adhesion molecule (204584_at), score: 0.54 LAMB2laminin, beta 2 (laminin S) (216264_s_at), score: 0.55 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: -0.52 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.65 LIMK1LIM domain kinase 1 (204357_s_at), score: -0.54 LMO7LIM domain 7 (202674_s_at), score: 0.55 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: -0.52 LOC51152melanoma antigen (220771_at), score: -0.53 LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: -0.6 LOC647070hypothetical LOC647070 (215467_x_at), score: -0.57 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.5 LPPR4plasticity related gene 1 (213496_at), score: 0.62 LSG1large subunit GTPase 1 homolog (S. cerevisiae) (221535_at), score: -0.49 LUMlumican (201744_s_at), score: 0.78 LXNlatexin (218729_at), score: 0.54 MAB21L1mab-21-like 1 (C. elegans) (206163_at), score: 0.53 MAGI2membrane associated guanylate kinase, WW and PDZ domain containing 2 (209737_at), score: 0.61 MANSC1MANSC domain containing 1 (220945_x_at), score: 0.55 MAP2K7mitogen-activated protein kinase kinase 7 (216206_x_at), score: -0.54 MBNL1muscleblind-like (Drosophila) (201151_s_at), score: 0.57 MED23mediator complex subunit 23 (218846_at), score: 0.56 MED31mediator complex subunit 31 (219318_x_at), score: -0.62 MEFVMediterranean fever (208262_x_at), score: -0.53 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: 0.57 METTL7Amethyltransferase like 7A (211424_x_at), score: -0.5 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: -0.54 MKRN2makorin ring finger protein 2 (218071_s_at), score: 0.54 MLF1myeloid leukemia factor 1 (204784_s_at), score: -0.52 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: 0.65 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: -0.53 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.56 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.91 MRPS18Cmitochondrial ribosomal protein S18C (220103_s_at), score: 0.56 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.56 MYL9myosin, light chain 9, regulatory (201058_s_at), score: 0.56 MYO1Bmyosin IB (212364_at), score: 0.74 NBEAneurobeachin (221207_s_at), score: 0.59 NCRNA00092non-protein coding RNA 92 (215861_at), score: -0.49 NDRG1N-myc downstream regulated 1 (200632_s_at), score: -0.54 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: 0.63 NESnestin (218678_at), score: 0.61 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: 0.54 NKIRAS2NFKB inhibitor interacting Ras-like 2 (218240_at), score: -0.49 NLGN1neuroligin 1 (205893_at), score: 0.59 NPAS2neuronal PAS domain protein 2 (39549_at), score: 0.59 NPTX1neuronal pentraxin I (204684_at), score: -0.63 NRN1neuritin 1 (218625_at), score: -0.52 NRP1neuropilin 1 (212298_at), score: 0.56 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: 0.56 OLIG2oligodendrocyte lineage transcription factor 2 (213825_at), score: -0.51 OR2A9Polfactory receptor, family 2, subfamily A, member 9 pseudogene (222290_at), score: 0.57 OSBPL11oxysterol binding protein-like 11 (218304_s_at), score: 0.54 OXTRoxytocin receptor (206825_at), score: 0.6 PAK2p21 protein (Cdc42/Rac)-activated kinase 2 (208878_s_at), score: 0.55 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: 0.61 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.77 PCOLCEprocollagen C-endopeptidase enhancer (202465_at), score: 0.53 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.54 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.72 PDLIM1PDZ and LIM domain 1 (208690_s_at), score: 0.65 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.54 PFDN2prefoldin subunit 2 (218336_at), score: -0.58 PGAP1post-GPI attachment to proteins 1 (213469_at), score: 0.54 PIM2pim-2 oncogene (204269_at), score: -0.51 PLAGL1pleiomorphic adenoma gene-like 1 (209318_x_at), score: 0.59 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.63 PLCG1phospholipase C, gamma 1 (202789_at), score: -0.58 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.5 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: -0.64 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: 0.56 PPP2R2Bprotein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (213849_s_at), score: -0.49 PRCCpapillary renal cell carcinoma (translocation-associated) (208938_at), score: -0.49 PRKCDBPprotein kinase C, delta binding protein (213010_at), score: 0.57 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: 0.55 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.65 PTPN13protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) (204201_s_at), score: 0.54 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.59 PYROXD1pyridine nucleotide-disulphide oxidoreductase domain 1 (213878_at), score: 0.56 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: 0.54 RARBretinoic acid receptor, beta (205080_at), score: -0.5 RASL11BRAS-like, family 11, member B (219142_at), score: 0.58 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: 0.58 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: -0.52 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.66 RND3Rho family GTPase 3 (212724_at), score: 0.59 ROD1ROD1 regulator of differentiation 1 (S. pombe) (214697_s_at), score: 0.53 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.58 RPL3ribosomal protein L3 (211666_x_at), score: -0.5 RPLP0P6ribosomal protein, large, P0 pseudogene 6 (214167_s_at), score: -0.51 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (218209_s_at), score: 0.53 RPS3ribosomal protein S3 (208692_at), score: -0.51 RPS4P17ribosomal protein S4X pseudogene 17 (216342_x_at), score: -0.53 RPS4Xribosomal protein S4, X-linked (200933_x_at), score: -0.53 RPS5ribosomal protein S5 (200024_at), score: -0.5 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: -0.53 RPS6KA2ribosomal protein S6 kinase, 90kDa, polypeptide 2 (212912_at), score: 0.57 S100A4S100 calcium binding protein A4 (203186_s_at), score: 0.56 SACSspastic ataxia of Charlevoix-Saguenay (sacsin) (213262_at), score: 0.58 SALL2sal-like 2 (Drosophila) (213283_s_at), score: 0.55 SCAND2SCAN domain containing 2 pseudogene (222177_s_at), score: -0.54 SCD5stearoyl-CoA desaturase 5 (220232_at), score: -0.5 SCRIBscribbled homolog (Drosophila) (212556_at), score: 0.57 SELTselenoprotein T (217811_at), score: 0.55 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.55 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.54 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.57 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.75 SFTPBsurfactant protein B (214354_x_at), score: -0.55 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: 0.63 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.5 SH3BGRSH3 domain binding glutamic acid-rich protein (204979_s_at), score: -0.57 SH3GL3SH3-domain GRB2-like 3 (211565_at), score: -0.66 SIM1single-minded homolog 1 (Drosophila) (206876_at), score: 0.55 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: 0.53 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.55 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: -0.54 SLC25A6solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 (212085_at), score: -0.52 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.62 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: -0.54 SLC35F2solute carrier family 35, member F2 (218826_at), score: -0.53 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.73 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: 0.58 SLC46A3solute carrier family 46, member 3 (214719_at), score: -0.49 SLFN12schlafen family member 12 (219885_at), score: 0.54 SMA4glucuronidase, beta pseudogene (215599_at), score: -0.64 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.64 SMAD4SMAD family member 4 (202527_s_at), score: 0.56 SNRPNsmall nuclear ribonucleoprotein polypeptide N (201522_x_at), score: 0.55 SOCS5suppressor of cytokine signaling 5 (209647_s_at), score: 0.58 SPANXCSPANX family, member C (220217_x_at), score: -0.66 SPATA20spermatogenesis associated 20 (218164_at), score: 0.56 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: -0.51 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: -0.59 SPNsialophorin (206057_x_at), score: -0.49 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: -0.53 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: 0.55 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: 0.54 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.57 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.57 STIM1stromal interaction molecule 1 (202764_at), score: -0.57 STOMstomatin (201060_x_at), score: 0.56 STSsteroid sulfatase (microsomal), isozyme S (203767_s_at), score: 0.57 STX12syntaxin 12 (212112_s_at), score: 0.53 SULF1sulfatase 1 (212353_at), score: 0.64 SULT1A1sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (203615_x_at), score: -0.49 SWAP70SWAP-70 protein (209307_at), score: 0.55 SYNGR1synaptogyrin 1 (210613_s_at), score: 0.63 TAGLNtransgelin (205547_s_at), score: 0.62 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: 0.53 TBXA2Rthromboxane A2 receptor (336_at), score: 0.75 TCF12transcription factor 12 (208986_at), score: 0.54 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: 0.61 TEStestis derived transcript (3 LIM domains) (202720_at), score: 0.7 TGFB2transforming growth factor, beta 2 (220407_s_at), score: -0.7 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: -0.56 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: -0.55 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: 0.54 THBS2thrombospondin 2 (203083_at), score: 0.61 TIGD1Ltigger transposable element derived 1-like (216459_x_at), score: -0.5 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: -0.55 TMEM2transmembrane protein 2 (218113_at), score: 0.57 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.58 TNFRSF9tumor necrosis factor receptor superfamily, member 9 (207536_s_at), score: -0.57 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: -0.52 TNS1tensin 1 (221748_s_at), score: 0.59 TOR3Atorsin family 3, member A (218459_at), score: -0.5 TPBGtrophoblast glycoprotein (203476_at), score: 0.58 TRAF5TNF receptor-associated factor 5 (204352_at), score: 0.55 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.63 TSKUtsukushin (218245_at), score: -0.48 TSPYL5TSPY-like 5 (213122_at), score: 0.61 TUBB6tubulin, beta 6 (209191_at), score: 0.57 TXNL4Bthioredoxin-like 4B (218794_s_at), score: -0.49 UBQLN4ubiquilin 4 (222252_x_at), score: -0.51 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: 0.57 UTP18UTP18, small subunit (SSU) processome component, homolog (yeast) (203721_s_at), score: -0.54 VEZTvezatin, adherens junctions transmembrane protein (207263_x_at), score: -0.52 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.69 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: 0.54 WDR44WD repeat domain 44 (219297_at), score: 0.53 WFDC8WAP four-disulfide core domain 8 (215276_at), score: -0.55 WWOXWW domain containing oxidoreductase (219077_s_at), score: -0.5 XRCC2X-ray repair complementing defective repair in Chinese hamster cells 2 (207598_x_at), score: -0.48 YTHDC2YTH domain containing 2 (213077_at), score: 0.56 ZBTB11zinc finger and BTB domain containing 11 (204847_at), score: 0.53 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: 0.53 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: -0.53 ZNF215zinc finger protein 215 (220214_at), score: 0.61 ZNF253zinc finger protein 253 (206900_x_at), score: -0.49 ZNF518Azinc finger protein 518A (204291_at), score: 0.6 ZNF580zinc finger protein 580 (220748_s_at), score: 0.54 ZNF675zinc finger protein 675 (217547_x_at), score: -0.49 ZNF816Azinc finger protein 816A (217541_x_at), score: -0.5
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |