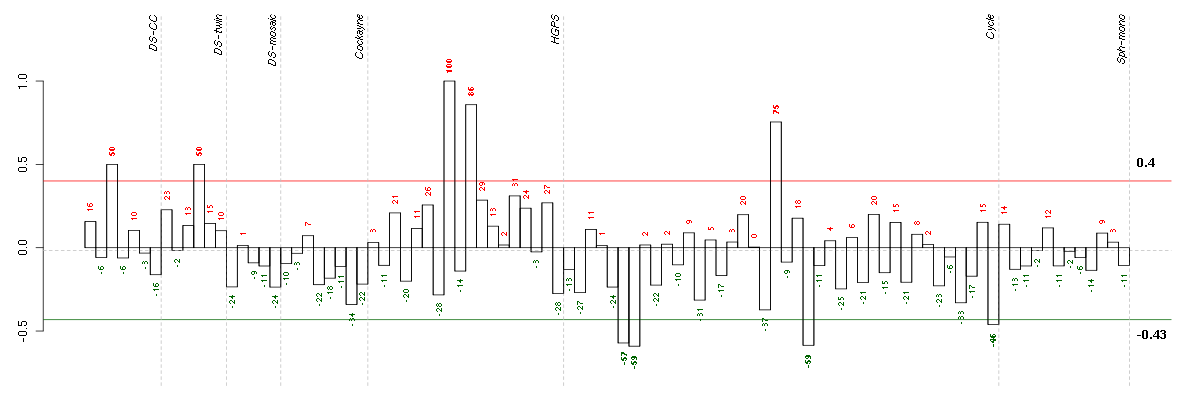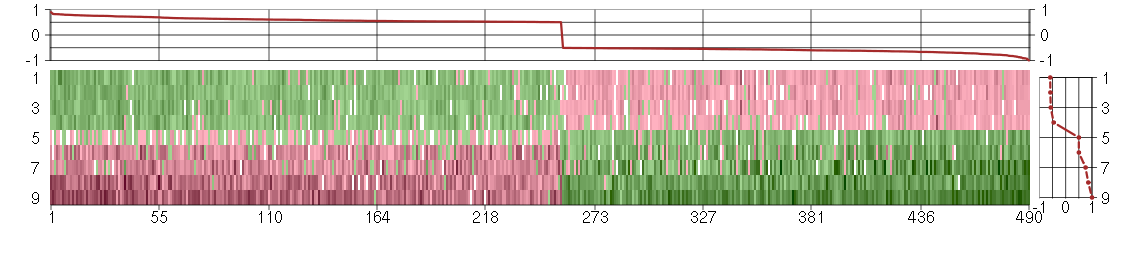

Under-expression is coded with green,
over-expression with red color.

vesicle-mediated transport
The directed movement of substances, either within a vesicle or in the vesicle membrane, into, out of or within a cell.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
intracellular transport
The directed movement of substances within a cell.
Golgi vesicle transport
The directed movement of substances into, out of or within the Golgi apparatus, mediated by vesicles.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
all
This term is the most general term possible
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
vesicle-mediated transport
The directed movement of substances, either within a vesicle or in the vesicle membrane, into, out of or within a cell.
intracellular transport
The directed movement of substances within a cell.
Golgi vesicle transport
The directed movement of substances into, out of or within the Golgi apparatus, mediated by vesicles.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
melanosome
A tissue-specific, membrane-bounded cytoplasmic organelle within which melanin pigments are synthesized and stored. Melanosomes are synthesized in melanocyte cells.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
pigment granule
A small, subcellular membrane-bounded vesicle containing pigment and/or pigment precursor molecules. Pigment granule biogenesis is poorly understood, as pigment granules are derived from multiple sources including the endoplasmic reticulum, coated vesicles, lysosomes, and endosomes.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.

ABHD11abhydrolase domain containing 11 (221927_s_at), score: 0.57 ABHD6abhydrolase domain containing 6 (221679_s_at), score: -0.52 ADAM10ADAM metallopeptidase domain 10 (214895_s_at), score: -0.7 ADAM9ADAM metallopeptidase domain 9 (meltrin gamma) (202381_at), score: -0.54 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.79 AFF4AF4/FMR2 family, member 4 (219199_at), score: -0.52 AHDC1AT hook, DNA binding motif, containing 1 (205002_at), score: 0.53 AHNAKAHNAK nucleoprotein (211986_at), score: 0.51 AKAP10A kinase (PRKA) anchor protein 10 (205045_at), score: -0.6 ANAPC2anaphase promoting complex subunit 2 (218555_at), score: 0.52 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: -0.57 API5apoptosis inhibitor 5 (214959_s_at), score: -0.55 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: 0.62 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (211975_at), score: 0.63 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: 0.53 ARHGEF4Rho guanine nucleotide exchange factor (GEF) 4 (205109_s_at), score: 0.53 ARL1ADP-ribosylation factor-like 1 (201657_at), score: -0.51 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: 0.7 ARL6IP5ADP-ribosylation-like factor 6 interacting protein 5 (200760_s_at), score: -0.6 ASF1AASF1 anti-silencing function 1 homolog A (S. cerevisiae) (203428_s_at), score: -0.61 ASPHaspartate beta-hydroxylase (205808_at), score: -0.54 ATN1atrophin 1 (40489_at), score: 0.77 ATP13A3ATPase type 13A3 (219558_at), score: -0.52 ATP6AP2ATPase, H+ transporting, lysosomal accessory protein 2 (201444_s_at), score: -0.51 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: -0.62 ATP6V1C1ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 (202874_s_at), score: -0.56 AUTS2autism susceptibility candidate 2 (212599_at), score: 0.56 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.78 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.76 BAIAP2BAI1-associated protein 2 (205294_at), score: 0.67 BCAT1branched chain aminotransferase 1, cytosolic (214452_at), score: -0.75 BCL2L1BCL2-like 1 (215037_s_at), score: -0.65 BCL7BB-cell CLL/lymphoma 7B (202518_at), score: 0.61 BCLAF1BCL2-associated transcription factor 1 (201101_s_at), score: -0.64 BIN1bridging integrator 1 (210201_x_at), score: 0.68 BMPR1Abone morphogenetic protein receptor, type IA (204832_s_at), score: -0.51 BRCC3BRCA1/BRCA2-containing complex, subunit 3 (216521_s_at), score: -0.64 BRD9bromodomain containing 9 (220155_s_at), score: 0.73 BRMS1breast cancer metastasis suppressor 1 (215631_s_at), score: 0.58 BST1bone marrow stromal cell antigen 1 (205715_at), score: -0.62 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.55 C10orf28chromosome 10 open reading frame 28 (210455_at), score: -0.57 C10orf72chromosome 10 open reading frame 72 (213381_at), score: 0.6 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.7 C18orf10chromosome 18 open reading frame 10 (213617_s_at), score: -0.51 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.51 C20orf30chromosome 20 open reading frame 30 (220477_s_at), score: -0.82 C2CD2LC2CD2-like (204757_s_at), score: 0.53 C3orf18chromosome 3 open reading frame 18 (219114_at), score: 0.53 C4orf8chromosome 4 open reading frame 8 (203600_s_at), score: 0.54 C5orf13chromosome 5 open reading frame 13 (201309_x_at), score: -0.61 C5orf28chromosome 5 open reading frame 28 (219029_at), score: -0.64 C6orf211chromosome 6 open reading frame 211 (218195_at), score: -0.51 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: -0.54 CA9carbonic anhydrase IX (205199_at), score: 0.53 CACNA2D1calcium channel, voltage-dependent, alpha 2/delta subunit 1 (207050_at), score: -0.53 CALD1caldesmon 1 (201616_s_at), score: -0.66 CALUcalumenin (214845_s_at), score: -0.62 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: 0.54 CANXcalnexin (208853_s_at), score: -0.72 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.6 CAPRIN1cell cycle associated protein 1 (200722_s_at), score: -0.72 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.53 CASKcalcium/calmodulin-dependent serine protein kinase (MAGUK family) (211208_s_at), score: 0.53 CCNE1cyclin E1 (213523_at), score: 0.6 CD164CD164 molecule, sialomucin (208653_s_at), score: -0.53 CD46CD46 molecule, complement regulatory protein (211574_s_at), score: -0.61 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.7 CDC42EP3CDC42 effector protein (Rho GTPase binding) 3 (209287_s_at), score: -0.57 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (209112_at), score: -0.54 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.79 CDV3CDV3 homolog (mouse) (213548_s_at), score: -0.65 CENPTcentromere protein T (218148_at), score: 0.65 CEP164centrosomal protein 164kDa (204251_s_at), score: 0.78 CHD8chromodomain helicase DNA binding protein 8 (212571_at), score: 0.64 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: 0.61 CICcapicua homolog (Drosophila) (212784_at), score: 0.68 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.56 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.57 CLDND1claudin domain containing 1 (208925_at), score: -0.52 CLGNcalmegin (205830_at), score: 0.6 CLIC4chloride intracellular channel 4 (201559_s_at), score: -0.94 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: -0.7 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.79 CNN3calponin 3, acidic (201445_at), score: -0.56 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.58 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.51 COL6A1collagen, type VI, alpha 1 (212940_at), score: -0.79 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: 0.67 COL8A1collagen, type VIII, alpha 1 (214587_at), score: -0.67 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.59 COMMD10COMM domain containing 10 (218439_s_at), score: -0.61 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.5 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: -0.66 CPDcarboxypeptidase D (201942_s_at), score: -0.63 CRIP1cysteine-rich protein 1 (intestinal) (205081_at), score: 0.57 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.64 CTDSPLCTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like (201904_s_at), score: 0.5 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.51 CYTH1cytohesin 1 (202880_s_at), score: 0.61 CYTH3cytohesin 3 (206523_at), score: -0.56 DAZAP2DAZ associated protein 2 (212595_s_at), score: -0.53 DCHS1dachsous 1 (Drosophila) (222101_s_at), score: 0.75 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.77 DDX5DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (200033_at), score: -0.61 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (219111_s_at), score: 0.82 DEGS1degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) (207431_s_at), score: -0.53 DENND4BDENN/MADD domain containing 4B (202860_at), score: 0.6 DGCR8DiGeorge syndrome critical region gene 8 (218650_at), score: 0.65 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.62 DMWDdystrophia myotonica, WD repeat containing (33768_at), score: 0.73 DNAJB2DnaJ (Hsp40) homolog, subfamily B, member 2 (202500_at), score: 0.52 DNAJB4DnaJ (Hsp40) homolog, subfamily B, member 4 (203811_s_at), score: -0.56 DNM1dynamin 1 (215116_s_at), score: 0.74 DNM3dynamin 3 (209839_at), score: 0.55 DPYDdihydropyrimidine dehydrogenase (204646_at), score: -0.53 DSTNdestrin (actin depolymerizing factor) (201021_s_at), score: -0.62 DULLARDdullard homolog (Xenopus laevis) (200035_at), score: 0.6 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: -0.78 EFEMP1EGF-containing fibulin-like extracellular matrix protein 1 (201842_s_at), score: -0.51 EGLN2egl nine homolog 2 (C. elegans) (220956_s_at), score: 0.52 EHMT1euchromatic histone-lysine N-methyltransferase 1 (219339_s_at), score: -0.53 EIF2S3eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa (205321_at), score: -0.79 EML4echinoderm microtubule associated protein like 4 (220386_s_at), score: -0.54 EMP2epithelial membrane protein 2 (204975_at), score: 0.52 ENO2enolase 2 (gamma, neuronal) (201313_at), score: 0.53 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: 0.53 ENSAendosulfine alpha (221487_s_at), score: -0.56 EPRSglutamyl-prolyl-tRNA synthetase (200841_s_at), score: -0.68 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: -0.63 EPS8L2EPS8-like 2 (218180_s_at), score: 0.51 ERLIN1ER lipid raft associated 1 (202444_s_at), score: -0.52 EXOC5exocyst complex component 5 (218748_s_at), score: -0.71 EZRezrin (208621_s_at), score: -0.66 F11coagulation factor XI (206610_s_at), score: -0.56 F2Rcoagulation factor II (thrombin) receptor (203989_x_at), score: -0.69 FAM108B1family with sequence similarity 108, member B1 (220285_at), score: -0.52 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: 0.56 FAM168Bfamily with sequence similarity 168, member B (212017_at), score: 0.72 FAM38Afamily with sequence similarity 38, member A (202771_at), score: 0.52 FAM3Afamily with sequence similarity 3, member A (38043_at), score: 0.73 FAM53Bfamily with sequence similarity 53, member B (203206_at), score: 0.52 FASNfatty acid synthase (212218_s_at), score: 0.57 FBN1fibrillin 1 (202765_s_at), score: -0.65 FBXL5F-box and leucine-rich repeat protein 5 (209004_s_at), score: -0.54 FBXO17F-box protein 17 (220233_at), score: 0.6 FBXO3F-box protein 3 (218432_at), score: -0.52 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.54 FHfumarate hydratase (203032_s_at), score: -0.77 FHITfragile histidine triad gene (206492_at), score: 0.54 FHL1four and a half LIM domains 1 (201539_s_at), score: -0.87 FHOD3formin homology 2 domain containing 3 (218980_at), score: 0.54 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.8 FN1fibronectin 1 (214701_s_at), score: -0.84 FNDC4fibronectin type III domain containing 4 (218843_at), score: 0.51 FNTAfarnesyltransferase, CAAX box, alpha (209471_s_at), score: -0.54 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: -0.51 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: -0.53 FOXK2forkhead box K2 (203064_s_at), score: 0.65 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.62 FXR1fragile X mental retardation, autosomal homolog 1 (201635_s_at), score: -0.53 GABRG3gamma-aminobutyric acid (GABA) A receptor, gamma 3 (216895_at), score: 0.51 GAKcyclin G associated kinase (202281_at), score: 0.53 GAS7growth arrest-specific 7 (202191_s_at), score: 0.71 GATCglutamyl-tRNA(Gln) amidotransferase, subunit C homolog (bacterial) (214711_at), score: -0.61 GDE1glycerophosphodiester phosphodiesterase 1 (202593_s_at), score: -0.62 GGNBP2gametogenetin binding protein 2 (218079_s_at), score: 0.56 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.65 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: -0.56 GNA13guanine nucleotide binding protein (G protein), alpha 13 (206917_at), score: -0.72 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: 0.55 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: -0.56 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: 0.71 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.53 GPD2glycerol-3-phosphate dehydrogenase 2 (mitochondrial) (210007_s_at), score: -0.58 GPR153G protein-coupled receptor 153 (221902_at), score: 0.5 GPR172AG protein-coupled receptor 172A (222155_s_at), score: 0.5 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.6 GTF2Igeneral transcription factor II, i (210892_s_at), score: -0.82 HDAC7histone deacetylase 7 (217937_s_at), score: 0.7 HECTD3HECT domain containing 3 (218632_at), score: 0.53 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: -0.54 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.75 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.54 HOXC10homeobox C10 (218959_at), score: 0.54 HOXC11homeobox C11 (206745_at), score: 0.66 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: -0.58 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.82 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: -0.56 HTATSF1HIV-1 Tat specific factor 1 (202601_s_at), score: -0.52 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.77 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: -0.61 IFNGR1interferon gamma receptor 1 (211676_s_at), score: -0.64 IL13RA1interleukin 13 receptor, alpha 1 (210904_s_at), score: -0.74 IL17RAinterleukin 17 receptor A (205707_at), score: 0.57 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (213804_at), score: 0.51 INPP5Einositol polyphosphate-5-phosphatase, 72 kDa (204706_at), score: 0.54 IPO8importin 8 (205701_at), score: -0.54 IQSEC1IQ motif and Sec7 domain 1 (203906_at), score: 0.65 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.62 ITGA5integrin, alpha 5 (fibronectin receptor, alpha polypeptide) (201389_at), score: 0.51 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: -0.53 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: 0.5 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: -0.65 KHSRPKH-type splicing regulatory protein (204372_s_at), score: 0.53 KIAA1199KIAA1199 (212942_s_at), score: -0.61 KIAA1467KIAA1467 (213234_at), score: 0.55 KITLGKIT ligand (211124_s_at), score: -0.58 KLC2kinesin light chain 2 (218906_x_at), score: 0.61 KLF9Kruppel-like factor 9 (203543_s_at), score: -0.54 KLHL20kelch-like 20 (Drosophila) (210635_s_at), score: -0.51 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.53 KRT18keratin 18 (201596_x_at), score: -0.52 KRT7keratin 7 (209016_s_at), score: -0.54 LAMB2laminin, beta 2 (laminin S) (216264_s_at), score: 0.53 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.53 LEPREL2leprecan-like 2 (204854_at), score: 0.52 LGALS8lectin, galactoside-binding, soluble, 8 (208934_s_at), score: -0.54 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: 0.57 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: -0.63 LMAN1lectin, mannose-binding, 1 (203294_s_at), score: -0.74 LMF2lipase maturation factor 2 (212682_s_at), score: 0.61 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.79 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.67 LOC100133166similar to neighbor of BRCA1 gene 1 (201383_s_at), score: -0.7 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.82 LOC399491LOC399491 protein (214035_x_at), score: 0.6 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.56 LOXlysyl oxidase (213640_s_at), score: -0.71 LOXL2lysyl oxidase-like 2 (202997_s_at), score: -0.58 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.61 LRFN4leucine rich repeat and fibronectin type III domain containing 4 (219491_at), score: 0.72 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200785_s_at), score: 0.65 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: 0.63 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.59 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.64 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (206750_at), score: -0.62 MANEAmannosidase, endo-alpha (219003_s_at), score: -0.52 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.62 MAP3K3mitogen-activated protein kinase kinase kinase 3 (203514_at), score: 0.54 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.51 MAP4K5mitogen-activated protein kinase kinase kinase kinase 5 (203553_s_at), score: -0.82 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.65 MAPK1mitogen-activated protein kinase 1 (208351_s_at), score: -0.58 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.56 MARCH7membrane-associated ring finger (C3HC4) 7 (202654_x_at), score: -0.59 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.75 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.53 MAXMYC associated factor X (210734_x_at), score: -0.75 MBD3methyl-CpG binding domain protein 3 (41160_at), score: 0.62 MBNL1muscleblind-like (Drosophila) (201151_s_at), score: -0.65 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: -0.91 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: -0.57 MED15mediator complex subunit 15 (222175_s_at), score: 0.55 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: -0.53 METT11D1methyltransferase 11 domain containing 1 (218366_x_at), score: 0.53 MFSD10major facilitator superfamily domain containing 10 (209215_at), score: 0.57 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: 0.52 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.66 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.71 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: 0.6 MORF4mortality factor 4 (221381_s_at), score: -0.54 MR1major histocompatibility complex, class I-related (207565_s_at), score: -0.67 MSL1male-specific lethal 1 homolog (Drosophila) (212708_at), score: 0.53 MSL2male-specific lethal 2 homolog (Drosophila) (218733_at), score: 0.55 MTA2metastasis associated 1 family, member 2 (203442_x_at), score: 0.72 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.51 MYO1Bmyosin IB (212364_at), score: -0.58 MYST3MYST histone acetyltransferase (monocytic leukemia) 3 (202423_at), score: 0.63 NAALAD2N-acetylated alpha-linked acidic dipeptidase 2 (222161_at), score: 0.64 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.9 NBPF1neuroblastoma breakpoint family, member 1 (212854_x_at), score: 0.52 NBPF12neuroblastoma breakpoint family, member 12 (213612_x_at), score: 0.55 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.59 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.53 NEFMneurofilament, medium polypeptide (205113_at), score: -0.55 NF2neurofibromin 2 (merlin) (211017_s_at), score: -0.55 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.63 NFYBnuclear transcription factor Y, beta (218129_s_at), score: -0.58 NKX3-1NK3 homeobox 1 (209706_at), score: 0.71 NMD3NMD3 homolog (S. cerevisiae) (218036_x_at), score: -0.54 NOL9nucleolar protein 9 (218754_at), score: 0.53 NOTCH2NLNotch homolog 2 (Drosophila) N-terminal like (214722_at), score: 0.65 NPHP4nephronophthisis 4 (213471_at), score: 0.63 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.75 NR1D1nuclear receptor subfamily 1, group D, member 1 (31637_s_at), score: 0.75 NRG1neuregulin 1 (206343_s_at), score: 0.52 NRXN3neurexin 3 (205795_at), score: -0.51 NUFIP1nuclear fragile X mental retardation protein interacting protein 1 (205134_s_at), score: 0.6 OLFM1olfactomedin 1 (213131_at), score: 0.75 OSMRoncostatin M receptor (205729_at), score: -0.67 OTUD3OTU domain containing 3 (213216_at), score: 0.75 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: -0.59 PAFAH1B2platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa (210160_at), score: -0.65 PAPOLApoly(A) polymerase alpha (212720_at), score: -0.59 PARVBparvin, beta (37965_at), score: 0.59 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.55 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.56 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.76 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.54 PDCD4programmed cell death 4 (neoplastic transformation inhibitor) (202731_at), score: -0.79 PDIA3protein disulfide isomerase family A, member 3 (208612_at), score: -0.58 PDLIM4PDZ and LIM domain 4 (211564_s_at), score: 0.57 PDLIM5PDZ and LIM domain 5 (216804_s_at), score: -0.57 PEX13peroxisomal biogenesis factor 13 (205246_at), score: -0.51 PEX26peroxisomal biogenesis factor 26 (219180_s_at), score: 0.65 PEX6peroxisomal biogenesis factor 6 (204545_at), score: 0.55 PGRMC1progesterone receptor membrane component 1 (201120_s_at), score: -0.6 PHLPPPH domain and leucine rich repeat protein phosphatase (212719_at), score: 0.69 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: -0.63 PICALMphosphatidylinositol binding clathrin assembly protein (215236_s_at), score: -0.66 PIGQphosphatidylinositol glycan anchor biosynthesis, class Q (204144_s_at), score: 0.52 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.78 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (219155_at), score: 0.52 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: 0.62 PKN1protein kinase N1 (202161_at), score: 0.63 PLCD1phospholipase C, delta 1 (205125_at), score: 0.58 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.51 PLXNA3plexin A3 (203623_at), score: 0.54 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.52 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: -0.5 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.64 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: 0.53 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.71 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.53 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.58 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.56 POSTNperiostin, osteoblast specific factor (210809_s_at), score: -0.62 PPM1Fprotein phosphatase 1F (PP2C domain containing) (203063_at), score: 0.6 PPP2R5Bprotein phosphatase 2, regulatory subunit B', beta isoform (635_s_at), score: 0.53 PQLC1PQ loop repeat containing 1 (218208_at), score: 0.59 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.54 PRKAR1Aprotein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) (200604_s_at), score: -0.55 PRPF4BPRP4 pre-mRNA processing factor 4 homolog B (yeast) (211090_s_at), score: -0.63 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: -0.51 PRR14proline rich 14 (218714_at), score: 0.55 PRRX1paired related homeobox 1 (205991_s_at), score: -0.68 PSEN1presenilin 1 (207782_s_at), score: -0.63 PTK7PTK7 protein tyrosine kinase 7 (207011_s_at), score: 0.6 PTPLAD1protein tyrosine phosphatase-like A domain containing 1 (217777_s_at), score: -0.54 PTPN18protein tyrosine phosphatase, non-receptor type 18 (brain-derived) (213521_at), score: 0.54 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.63 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: -0.57 PXMP3peroxisomal membrane protein 3, 35kDa (210296_s_at), score: -0.6 RAB11FIP5RAB11 family interacting protein 5 (class I) (210879_s_at), score: 0.72 RAB15RAB15, member RAS onocogene family (221810_at), score: 0.72 RAB28RAB28, member RAS oncogene family (207495_at), score: -0.56 RAB2ARAB2A, member RAS oncogene family (208734_x_at), score: -0.52 RAB35RAB35, member RAS oncogene family (205461_at), score: -0.69 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: -1 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.73 RBBP4retinoblastoma binding protein 4 (217301_x_at), score: -0.54 RBBP9retinoblastoma binding protein 9 (221440_s_at), score: -0.61 RBM10RNA binding motif protein 10 (215089_s_at), score: 0.52 RBM15BRNA binding motif protein 15B (202689_at), score: 0.64 RBM9RNA binding motif protein 9 (213901_x_at), score: -0.51 RCAN1regulator of calcineurin 1 (215253_s_at), score: -0.64 RDXradixin (204969_s_at), score: -0.77 RELNreelin (205923_at), score: -0.51 RNF11ring finger protein 11 (208924_at), score: -0.51 RNF128ring finger protein 128 (219263_at), score: 0.55 RNF187ring finger protein 187 (212155_at), score: 0.68 RNF19Bring finger protein 19B (213038_at), score: 0.54 RNF220ring finger protein 220 (219988_s_at), score: 0.75 RNF24ring finger protein 24 (204669_s_at), score: 0.55 RNF6ring finger protein (C3H2C3 type) 6 (210932_s_at), score: -0.52 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: -0.64 RNGTTRNA guanylyltransferase and 5'-phosphatase (204207_s_at), score: -0.52 RPEribulose-5-phosphate-3-epimerase (216574_s_at), score: -0.6 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.53 RRN3RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) (216902_s_at), score: -0.51 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.52 RSL24D1ribosomal L24 domain containing 1 (217915_s_at), score: -0.63 S100A2S100 calcium binding protein A2 (204268_at), score: 0.54 SATB2SATB homeobox 2 (213435_at), score: 0.61 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.53 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: -0.85 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.74 SCARB2scavenger receptor class B, member 2 (201647_s_at), score: -0.62 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: -0.61 SCP2sterol carrier protein 2 (201339_s_at), score: -0.6 SCRN3secernin 3 (219234_x_at), score: -0.6 SDC1syndecan 1 (201287_s_at), score: 0.52 SDC2syndecan 2 (212154_at), score: -0.52 SDC4syndecan 4 (202071_at), score: 0.6 SEC22BSEC22 vesicle trafficking protein homolog B (S. cerevisiae) (214257_s_at), score: -0.55 SEC23ASec23 homolog A (S. cerevisiae) (204344_s_at), score: -0.71 SELTselenoprotein T (217811_at), score: -0.55 SEPT11septin 11 (201308_s_at), score: -0.62 SETBP1SET binding protein 1 (205933_at), score: 0.74 SF1splicing factor 1 (208313_s_at), score: 0.66 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.71 SFRS1splicing factor, arginine/serine-rich 1 (201742_x_at), score: -0.57 SFRS6splicing factor, arginine/serine-rich 6 (206108_s_at), score: -0.52 SGCBsarcoglycan, beta (43kDa dystrophin-associated glycoprotein) (205121_at), score: -0.52 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.56 SIPA1L3signal-induced proliferation-associated 1 like 3 (213600_at), score: 0.77 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (203727_at), score: 0.55 SLC25A21solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21 (220474_at), score: 0.5 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.52 SLC6A8solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (202219_at), score: 0.75 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: 0.58 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (212947_at), score: 0.69 SLCO3A1solute carrier organic anion transporter family, member 3A1 (219229_at), score: 0.55 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (215294_s_at), score: -0.52 SMARCA4SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 (212520_s_at), score: 0.52 SMARCD1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 (209518_at), score: 0.51 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.58 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.73 SMOXspermine oxidase (210357_s_at), score: 0.51 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: -0.91 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.67 SPAG9sperm associated antigen 9 (206748_s_at), score: -0.56 SPON1spondin 1, extracellular matrix protein (209436_at), score: 0.72 SPTLC1serine palmitoyltransferase, long chain base subunit 1 (202278_s_at), score: -0.58 SRGNserglycin (201859_at), score: -0.61 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: -0.75 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: -0.61 STARD8StAR-related lipid transfer (START) domain containing 8 (206868_at), score: 0.56 STAT1signal transducer and activator of transcription 1, 91kDa (AFFX-HUMISGF3A/M97935_MA_at), score: -0.68 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: -0.51 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: -0.64 STOMstomatin (201060_x_at), score: -0.55 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.82 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: 0.62 SUSD5sushi domain containing 5 (214954_at), score: -0.56 SUV39H1suppressor of variegation 3-9 homolog 1 (Drosophila) (218619_s_at), score: 0.53 SYPL1synaptophysin-like 1 (201259_s_at), score: -0.57 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: -0.72 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.6 TATDN2TatD DNase domain containing 2 (203648_at), score: 0.65 TBC1D10BTBC1 domain family, member 10B (220947_s_at), score: 0.62 TBCDtubulin folding cofactor D (211052_s_at), score: 0.66 TBL1XR1transducin (beta)-like 1 X-linked receptor 1 (221428_s_at), score: -0.55 TBL3transducin (beta)-like 3 (209820_s_at), score: 0.62 TBX3T-box 3 (219682_s_at), score: 0.53 TCF20transcription factor 20 (AR1) (212931_at), score: 0.61 TCF7transcription factor 7 (T-cell specific, HMG-box) (205255_x_at), score: 0.66 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (34260_at), score: 0.61 TFPItissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) (209676_at), score: -0.66 TGFAtransforming growth factor, alpha (205016_at), score: 0.63 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.54 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: -0.57 TJP1tight junction protein 1 (zona occludens 1) (214168_s_at), score: -0.55 TM4SF1transmembrane 4 L six family member 1 (215034_s_at), score: -0.52 TMED2transmembrane emp24 domain trafficking protein 2 (204426_at), score: -0.88 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.7 TMEM123transmembrane protein 123 (211967_at), score: -0.59 TMEM158transmembrane protein 158 (213338_at), score: 0.62 TMOD3tropomodulin 3 (ubiquitous) (220800_s_at), score: -0.59 TMX1thioredoxin-related transmembrane protein 1 (208097_s_at), score: -0.63 TNFRSF10Dtumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (210654_at), score: -0.69 TNFRSF11Btumor necrosis factor receptor superfamily, member 11b (204933_s_at), score: -0.54 TNK2tyrosine kinase, non-receptor, 2 (203839_s_at), score: 0.56 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.56 TPMTthiopurine S-methyltransferase (203672_x_at), score: -0.54 TRAM1translocation associated membrane protein 1 (210733_at), score: -0.53 TRIM33tripartite motif-containing 33 (210266_s_at), score: 0.57 TRMT11tRNA methyltransferase 11 homolog (S. cerevisiae) (218877_s_at), score: -0.54 TSPAN3tetraspanin 3 (200973_s_at), score: -0.59 TWF1twinfilin, actin-binding protein, homolog 1 (Drosophila) (214007_s_at), score: -0.66 TXLNAtaxilin alpha (212300_at), score: 0.69 TXNRD1thioredoxin reductase 1 (201266_at), score: -0.55 UBE2D3ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) (200669_s_at), score: -0.54 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: -0.6 UBE2Oubiquitin-conjugating enzyme E2O (218141_at), score: 0.59 UBE2Zubiquitin-conjugating enzyme E2Z (217750_s_at), score: 0.54 UBXN4UBX domain protein 4 (212008_at), score: -0.72 USP34ubiquitin specific peptidase 34 (212065_s_at), score: -0.61 UTRNutrophin (213022_s_at), score: -0.69 VAMP3vesicle-associated membrane protein 3 (cellubrevin) (201337_s_at), score: -0.68 VCANversican (215646_s_at), score: -0.53 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.76 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: -0.67 WDR1WD repeat domain 1 (210935_s_at), score: -0.76 WDR62WD repeat domain 62 (215218_s_at), score: 0.56 WRAP53WD repeat containing, antisense to TP53 (44563_at), score: 0.54 XAB2XPA binding protein 2 (218110_at), score: 0.78 YDD19YDD19 protein (37079_at), score: -0.51 YIPF5Yip1 domain family, member 5 (221423_s_at), score: -0.62 YWHAZtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (200641_s_at), score: -0.63 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: 0.62 ZFAND5zinc finger, AN1-type domain 5 (217741_s_at), score: -0.77 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.53 ZMYND11zinc finger, MYND domain containing 11 (202137_s_at), score: -0.56 ZNF236zinc finger protein 236 (47571_at), score: 0.53 ZNF318zinc finger protein 318 (203521_s_at), score: 0.55 ZNF536zinc finger protein 536 (206403_at), score: 0.8 ZNF675zinc finger protein 675 (217547_x_at), score: -0.51 ZNF692zinc finger protein 692 (220661_s_at), score: 0.52
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |