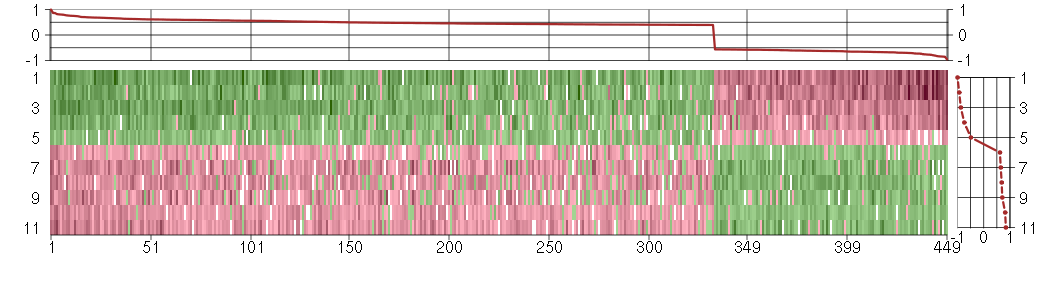

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
carbohydrate metabolic process
The chemical reactions and pathways involving carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y. Includes the formation of carbohydrate derivatives by the addition of a carbohydrate residue to another molecule.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
cellular carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, carried out by individual cells.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular carbohydrate metabolic process
The chemical reactions and pathways involving carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, as carried out by individual cells.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, carried out by individual cells.
carbohydrate biosynthetic process
The chemical reactions and pathways resulting in the formation of carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y.
cellular carbohydrate metabolic process
The chemical reactions and pathways involving carbohydrates, any of a group of organic compounds based of the general formula Cx(H2O)y, as carried out by individual cells.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -0.96 ABHD14Aabhydrolase domain containing 14A (210006_at), score: 0.54 ACP2acid phosphatase 2, lysosomal (202767_at), score: 0.47 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: 0.58 ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: 0.42 ADCY7adenylate cyclase 7 (203741_s_at), score: 0.49 ADORA1adenosine A1 receptor (216220_s_at), score: 0.56 ADORA2Badenosine A2b receptor (205891_at), score: -0.62 AHNAK2AHNAK nucleoprotein 2 (212992_at), score: 0.48 AIM2absent in melanoma 2 (206513_at), score: -0.68 ALDH1B1aldehyde dehydrogenase 1 family, member B1 (209646_x_at), score: 0.45 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: 0.44 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: -0.73 ANKRD6ankyrin repeat domain 6 (204671_s_at), score: -0.61 ANPEPalanyl (membrane) aminopeptidase (202888_s_at), score: 0.47 ANTXR1anthrax toxin receptor 1 (220092_s_at), score: 0.56 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.73 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (217888_s_at), score: 0.45 ARHGAP1Rho GTPase activating protein 1 (202117_at), score: 0.4 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: 0.46 ARSAarylsulfatase A (204443_at), score: 0.51 ATF5activating transcription factor 5 (204999_s_at), score: -0.57 ATP6V0CATPase, H+ transporting, lysosomal 16kDa, V0 subunit c (36994_at), score: 0.43 ATP8B2ATPase, class I, type 8B, member 2 (216873_s_at), score: 0.4 ATP9AATPase, class II, type 9A (212062_at), score: 0.46 AUTS2autism susceptibility candidate 2 (212599_at), score: 0.4 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: 0.46 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: 0.7 BACE1beta-site APP-cleaving enzyme 1 (217904_s_at), score: 0.55 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.5 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: -0.67 BCKDKbranched chain ketoacid dehydrogenase kinase (202030_at), score: 0.5 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: -0.63 BSCL2Bernardinelli-Seip congenital lipodystrophy 2 (seipin) (208906_at), score: 0.4 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.57 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: 0.41 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.4 C19orf10chromosome 19 open reading frame 10 (216483_s_at), score: 0.41 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.55 C20orf39chromosome 20 open reading frame 39 (219310_at), score: 0.47 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.66 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.86 C5orf23chromosome 5 open reading frame 23 (219054_at), score: 0.69 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.75 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: 0.48 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.67 CAPN1calpain 1, (mu/I) large subunit (200752_s_at), score: 0.4 CARD10caspase recruitment domain family, member 10 (210026_s_at), score: -0.71 CCND2cyclin D2 (200953_s_at), score: -0.64 CD248CD248 molecule, endosialin (219025_at), score: 0.52 CD68CD68 molecule (203507_at), score: 0.46 CD81CD81 molecule (200675_at), score: 0.47 CDCP1CUB domain containing protein 1 (218451_at), score: 0.4 CDH4cadherin 4, type 1, R-cadherin (retinal) (220227_at), score: 0.52 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.41 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.66 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 0.82 CH25Hcholesterol 25-hydroxylase (206932_at), score: 0.46 CHEK2CHK2 checkpoint homolog (S. pombe) (210416_s_at), score: -0.57 CHN1chimerin (chimaerin) 1 (212624_s_at), score: 0.44 CHPFchondroitin polymerizing factor (202175_at), score: 0.63 CHST12carbohydrate (chondroitin 4) sulfotransferase 12 (218927_s_at), score: 0.43 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: 0.47 CIB1calcium and integrin binding 1 (calmyrin) (201953_at), score: 0.44 CKBcreatine kinase, brain (200884_at), score: 0.63 CLDND1claudin domain containing 1 (208925_at), score: -0.6 CLIC3chloride intracellular channel 3 (219529_at), score: 0.47 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.48 COL11A1collagen, type XI, alpha 1 (37892_at), score: 0.51 COL13A1collagen, type XIII, alpha 1 (211343_s_at), score: 0.43 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.69 COMPcartilage oligomeric matrix protein (205713_s_at), score: 1 COPGcoatomer protein complex, subunit gamma (217749_at), score: 0.4 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: 0.54 CRABP2cellular retinoic acid binding protein 2 (202575_at), score: 0.57 CRATcarnitine acetyltransferase (209522_s_at), score: 0.6 CREB3L1cAMP responsive element binding protein 3-like 1 (213059_at), score: 0.42 CRLF1cytokine receptor-like factor 1 (206315_at), score: 0.4 CRTAPcartilage associated protein (201380_at), score: 0.42 CRY1cryptochrome 1 (photolyase-like) (209674_at), score: -0.65 CRYABcrystallin, alpha B (209283_at), score: 0.52 CSDC2cold shock domain containing C2, RNA binding (209981_at), score: 0.4 CST6cystatin E/M (206595_at), score: 0.61 CTSCcathepsin C (201487_at), score: -0.67 CTSKcathepsin K (202450_s_at), score: 0.41 CYBAcytochrome b-245, alpha polypeptide (203028_s_at), score: 0.48 CYFIP1cytoplasmic FMR1 interacting protein 1 (208923_at), score: 0.41 CYP1B1cytochrome P450, family 1, subfamily B, polypeptide 1 (202436_s_at), score: -0.57 DBF4DBF4 homolog (S. cerevisiae) (204244_s_at), score: -0.58 DCHS1dachsous 1 (Drosophila) (222101_s_at), score: 0.44 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.67 DDX28DEAD (Asp-Glu-Ala-Asp) box polypeptide 28 (203785_s_at), score: -0.57 DDX5DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (200033_at), score: -0.57 DENND5BDENN/MADD domain containing 5B (215058_at), score: 0.41 DNM1dynamin 1 (215116_s_at), score: 0.46 DPYSL4dihydropyrimidinase-like 4 (205493_s_at), score: 0.44 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.62 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.59 ECOPEGFR-coamplified and overexpressed protein (208091_s_at), score: 0.42 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.42 EFEMP2EGF-containing fibulin-like extracellular matrix protein 2 (209356_x_at), score: 0.45 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: 0.56 ELL2elongation factor, RNA polymerase II, 2 (214446_at), score: 0.44 EMILIN1elastin microfibril interfacer 1 (204163_at), score: 0.41 EMP3epithelial membrane protein 3 (203729_at), score: 0.41 EMX2empty spiracles homeobox 2 (221950_at), score: 0.52 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: 0.62 EPB41L3erythrocyte membrane protein band 4.1-like 3 (206710_s_at), score: 0.6 EPS8L2EPS8-like 2 (218180_s_at), score: 0.47 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: 0.49 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: 0.57 FAM155Afamily with sequence similarity 155, member A (214825_at), score: 0.44 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.69 FAM46Afamily with sequence similarity 46, member A (221766_s_at), score: 0.53 FASFas (TNF receptor superfamily, member 6) (204780_s_at), score: 0.58 FASNfatty acid synthase (212218_s_at), score: 0.49 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.51 FBLN5fibulin 5 (203088_at), score: 0.43 FBN2fibrillin 2 (203184_at), score: 0.45 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: 0.59 FGF2fibroblast growth factor 2 (basic) (204421_s_at), score: 0.43 FHOD3formin homology 2 domain containing 3 (218980_at), score: 0.69 FKBP4FK506 binding protein 4, 59kDa (200894_s_at), score: -0.61 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.43 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.56 FLJ11151hypothetical protein FLJ11151 (218610_s_at), score: 0.41 FLNCfilamin C, gamma (actin binding protein 280) (207876_s_at), score: 0.49 FLRT2fibronectin leucine rich transmembrane protein 2 (204359_at), score: 0.55 FNDC4fibronectin type III domain containing 4 (218843_at), score: 0.41 FOLR3folate receptor 3 (gamma) (206371_at), score: 0.59 FOXD1forkhead box D1 (206307_s_at), score: 0.49 FXYD5FXYD domain containing ion transport regulator 5 (218084_x_at), score: 0.46 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.71 GABRA5gamma-aminobutyric acid (GABA) A receptor, alpha 5 (206456_at), score: 0.42 GAGE4G antigen 4 (207086_x_at), score: -0.8 GAGE6G antigen 6 (208155_x_at), score: -0.83 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (219956_at), score: 0.49 GAS1growth arrest-specific 1 (204457_s_at), score: 0.4 GAS6growth arrest-specific 6 (202177_at), score: 0.68 GATA3GATA binding protein 3 (209604_s_at), score: -0.66 GATA6GATA binding protein 6 (210002_at), score: -0.65 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (202269_x_at), score: -0.7 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.45 GLSglutaminase (203159_at), score: 0.61 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.46 GLTPglycolipid transfer protein (219267_at), score: 0.49 GMDSGDP-mannose 4,6-dehydratase (204875_s_at), score: -0.63 GNAI1guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 (209576_at), score: -0.57 GPAA1glycosylphosphatidylinositol anchor attachment protein 1 homolog (yeast) (215690_x_at), score: 0.53 GPERG protein-coupled estrogen receptor 1 (210640_s_at), score: 0.42 GPR124G protein-coupled receptor 124 (221814_at), score: 0.4 GPR89AG protein-coupled receptor 89A (222140_s_at), score: 0.43 GPR89BG protein-coupled receptor 89B (220642_x_at), score: 0.4 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.45 GREM2gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) (220794_at), score: 0.44 GSRglutathione reductase (205770_at), score: -0.58 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: 0.6 HAS2hyaluronan synthase 2 (206432_at), score: 0.63 HEPHhephaestin (203903_s_at), score: 0.43 HERC5hect domain and RLD 5 (219863_at), score: -0.67 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.48 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: -0.61 HIST1H4Chistone cluster 1, H4c (205967_at), score: 0.55 HIST2H2AA3histone cluster 2, H2aa3 (214290_s_at), score: 0.67 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.56 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.65 HOXA10homeobox A10 (213150_at), score: 0.41 HOXA9homeobox A9 (214651_s_at), score: 0.4 HOXB2homeobox B2 (205453_at), score: 0.5 HOXD4homeobox D4 (205522_at), score: 0.41 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: 0.56 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.65 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.47 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.56 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: 0.8 IDSiduronate 2-sulfatase (202439_s_at), score: -0.57 IER3immediate early response 3 (201631_s_at), score: -0.63 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: -0.77 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: -0.58 IGFBP3insulin-like growth factor binding protein 3 (212143_s_at), score: 0.6 IL11interleukin 11 (206924_at), score: 0.4 IL11RAinterleukin 11 receptor, alpha (204773_at), score: 0.54 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.51 INHBBinhibin, beta B (205258_at), score: 0.55 IPO8importin 8 (205701_at), score: -0.57 IQSEC1IQ motif and Sec7 domain 1 (203906_at), score: 0.44 IRF6interferon regulatory factor 6 (202597_at), score: 0.47 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.66 IRX5iroquois homeobox 5 (210239_at), score: 0.74 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.86 JARID1Djumonji, AT rich interactive domain 1D (206700_s_at), score: 0.4 KBTBD11kelch repeat and BTB (POZ) domain containing 11 (204301_at), score: 0.47 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: -0.61 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: 0.56 KIAA0100KIAA0100 (201729_s_at), score: 0.4 KIAA0495KIAA0495 (213340_s_at), score: 0.6 KIAA1539KIAA1539 (207765_s_at), score: 0.49 KIF1Ckinesin family member 1C (209244_s_at), score: 0.44 KIF3Akinesin family member 3A (213623_at), score: -0.61 KLF9Kruppel-like factor 9 (203543_s_at), score: -0.59 KRT14keratin 14 (209351_at), score: 0.63 KRT19keratin 19 (201650_at), score: 0.6 KRT34keratin 34 (206969_at), score: 0.48 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: 0.47 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.73 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: 0.42 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.63 LMF1lipase maturation factor 1 (219135_s_at), score: 0.51 LOC151162hypothetical LOC151162 (212098_at), score: 0.48 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.77 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: 0.41 LRFN4leucine rich repeat and fibronectin type III domain containing 4 (219491_at), score: 0.4 LRP10low density lipoprotein receptor-related protein 10 (201412_at), score: 0.44 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: 0.44 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.69 LRRFIP1leucine rich repeat (in FLII) interacting protein 1 (201861_s_at), score: 0.59 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.57 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: -0.68 MALLmal, T-cell differentiation protein-like (209373_at), score: 0.61 MALT1mucosa associated lymphoid tissue lymphoma translocation gene 1 (208309_s_at), score: 0.4 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: 0.43 MAP3K3mitogen-activated protein kinase kinase kinase 3 (203514_at), score: 0.44 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.85 ME1malic enzyme 1, NADP(+)-dependent, cytosolic (204059_s_at), score: -0.66 MEGF6multiple EGF-like-domains 6 (213942_at), score: 0.63 MFAP5microfibrillar associated protein 5 (213764_s_at), score: 0.59 MFSD10major facilitator superfamily domain containing 10 (209215_at), score: 0.42 MICBMHC class I polypeptide-related sequence B (206247_at), score: -0.62 MITFmicrophthalmia-associated transcription factor (207233_s_at), score: -0.76 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: 0.56 MMP17matrix metallopeptidase 17 (membrane-inserted) (206234_s_at), score: 0.39 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: 0.49 MMP3matrix metallopeptidase 3 (stromelysin 1, progelatinase) (205828_at), score: 0.43 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.55 MRASmuscle RAS oncogene homolog (206538_at), score: 0.48 MRC2mannose receptor, C type 2 (37408_at), score: 0.51 MRFAP1L1Morf4 family associated protein 1-like 1 (212199_at), score: -0.58 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: -0.7 MSL2male-specific lethal 2 homolog (Drosophila) (218733_at), score: 0.48 MSX1msh homeobox 1 (205932_s_at), score: 0.52 MT1Gmetallothionein 1G (204745_x_at), score: 0.51 MT1Mmetallothionein 1M (217546_at), score: 0.74 MT1P3metallothionein 1 pseudogene 3 (221953_s_at), score: 0.59 MT1Xmetallothionein 1X (204326_x_at), score: 0.58 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.68 MXRA7matrix-remodelling associated 7 (212509_s_at), score: 0.4 MXRA8matrix-remodelling associated 8 (213422_s_at), score: 0.6 MYBL1v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (213906_at), score: 0.41 MYO10myosin X (201976_s_at), score: 0.52 MYO1Cmyosin IC (214656_x_at), score: 0.62 NAAAN-acylethanolamine acid amidase (214765_s_at), score: 0.43 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: -0.58 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.65 NCALDneurocalcin delta (211685_s_at), score: 0.57 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.43 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: 0.44 NDNnecdin homolog (mouse) (209550_at), score: 0.7 NDRG3NDRG family member 3 (217286_s_at), score: 0.47 NDST1N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (202607_at), score: 0.53 NEFLneurofilament, light polypeptide (221805_at), score: -0.7 NEK11NIMA (never in mitosis gene a)- related kinase 11 (219542_at), score: 0.43 NET1neuroepithelial cell transforming 1 (201830_s_at), score: 0.46 NFASCneurofascin homolog (chicken) (213438_at), score: 0.52 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (208003_s_at), score: -0.58 NFYBnuclear transcription factor Y, beta (218129_s_at), score: -0.64 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: -0.6 NID2nidogen 2 (osteonidogen) (204114_at), score: -0.59 NMIN-myc (and STAT) interactor (203964_at), score: -0.62 NMUneuromedin U (206023_at), score: -0.65 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.47 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: 0.52 NPTX2neuronal pentraxin II (213479_at), score: 0.48 NR2F2nuclear receptor subfamily 2, group F, member 2 (215073_s_at), score: -0.57 NRG1neuregulin 1 (206343_s_at), score: 0.59 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.63 NRN1neuritin 1 (218625_at), score: 0.4 NTF3neurotrophin 3 (206706_at), score: 0.61 NTMneurotrimin (222020_s_at), score: 0.43 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.84 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: 0.61 OLFML3olfactomedin-like 3 (218162_at), score: 0.41 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.41 OSBPL10oxysterol binding protein-like 10 (219073_s_at), score: -0.58 OXTRoxytocin receptor (206825_at), score: 0.4 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: 0.5 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: 0.46 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (222125_s_at), score: 0.43 PACS1phosphofurin acidic cluster sorting protein 1 (220557_s_at), score: 0.41 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: -0.7 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.68 PCBP4poly(rC) binding protein 4 (209361_s_at), score: 0.52 PCOLCE2procollagen C-endopeptidase enhancer 2 (219295_s_at), score: 0.39 PDE10Aphosphodiesterase 10A (205501_at), score: -0.84 PDGFCplatelet derived growth factor C (218718_at), score: 0.42 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: 0.53 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: -0.63 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.7 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.41 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: -0.63 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (219155_at), score: 0.59 PLA2G15phospholipase A2, group XV (204458_at), score: 0.47 PLA2G16phospholipase A2, group XVI (209581_at), score: -0.61 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (210145_at), score: -0.67 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.42 PLXNA3plexin A3 (203623_at), score: 0.42 PNMA2paraneoplastic antigen MA2 (209598_at), score: -0.81 PODXLpodocalyxin-like (201578_at), score: 0.67 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.57 POMT1protein-O-mannosyltransferase 1 (218476_at), score: 0.43 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.46 PPAP2Aphosphatidic acid phosphatase type 2A (209147_s_at), score: 0.44 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: -0.58 PPP1CBprotein phosphatase 1, catalytic subunit, beta isoform (201409_s_at), score: 0.43 PPP1R13Lprotein phosphatase 1, regulatory (inhibitor) subunit 13 like (218849_s_at), score: 0.45 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: 0.65 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.43 PRKCHprotein kinase C, eta (218764_at), score: -0.68 PRPF38BPRP38 pre-mRNA processing factor 38 (yeast) domain containing B (218040_at), score: -0.61 PRPS1phosphoribosyl pyrophosphate synthetase 1 (209440_at), score: 0.47 PSD4pleckstrin and Sec7 domain containing 4 (203317_at), score: 0.49 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: 0.56 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: 0.59 PSG6pregnancy specific beta-1-glycoprotein 6 (209738_x_at), score: 0.52 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.62 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (202243_s_at), score: 0.46 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.62 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: -0.61 PTGER3prostaglandin E receptor 3 (subtype EP3) (213933_at), score: 0.58 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: 0.43 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.39 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: 0.77 PTPRRprotein tyrosine phosphatase, receptor type, R (210675_s_at), score: 0.43 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.45 R3HDM2R3H domain containing 2 (203831_at), score: 0.41 RAB40BRAB40B, member RAS oncogene family (204547_at), score: 0.48 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: -0.67 RAD23BRAD23 homolog B (S. cerevisiae) (201223_s_at), score: 0.55 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: -0.58 RASA4RAS p21 protein activator 4 (208534_s_at), score: 0.42 RASIP1Ras interacting protein 1 (220027_s_at), score: 0.63 RBM12BRNA binding motif protein 12B (51228_at), score: -0.63 RBM3RNA binding motif (RNP1, RRM) protein 3 (222026_at), score: 0.59 RBM39RNA binding motif protein 39 (207941_s_at), score: -0.57 RCN3reticulocalbin 3, EF-hand calcium binding domain (61734_at), score: 0.47 RDXradixin (204969_s_at), score: -0.57 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (212968_at), score: 0.42 RGS7regulator of G-protein signaling 7 (206290_s_at), score: -0.6 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (219489_s_at), score: 0.4 RHODras homolog gene family, member D (31846_at), score: 0.44 RNF128ring finger protein 128 (219263_at), score: 0.44 RORARAR-related orphan receptor A (210426_x_at), score: 0.44 RP6-213H19.1serine/threonine protein kinase MST4 (218499_at), score: -0.6 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.4 RTN1reticulon 1 (203485_at), score: -0.71 RTN3reticulon 3 (219549_s_at), score: 0.53 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.56 SAP18Sin3A-associated protein, 18kDa (208740_at), score: 0.43 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: -0.57 SCG5secretogranin V (7B2 protein) (203889_at), score: 0.56 SDC1syndecan 1 (201287_s_at), score: 0.4 SDC4syndecan 4 (202071_at), score: 0.49 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.42 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.62 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.81 SH3PXD2ASH3 and PX domains 2A (213252_at), score: 0.39 SIRPAsignal-regulatory protein alpha (202897_at), score: 0.55 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: 0.53 SLC12A2solute carrier family 12 (sodium/potassium/chloride transporters), member 2 (204404_at), score: -0.62 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: 0.58 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.51 SLC17A9solute carrier family 17, member 9 (219559_at), score: 0.45 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.57 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: 0.49 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.57 SLC39A4solute carrier family 39 (zinc transporter), member 4 (219215_s_at), score: 0.4 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: 0.68 SLC7A1solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 (212295_s_at), score: 0.61 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: -0.85 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: 0.6 SMAD3SMAD family member 3 (218284_at), score: -0.67 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.62 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: 0.57 SNAI1snail homolog 1 (Drosophila) (219480_at), score: 0.46 SNAPC1small nuclear RNA activating complex, polypeptide 1, 43kDa (205443_at), score: -0.57 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.65 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (208608_s_at), score: -0.58 SOBPsine oculis binding protein homolog (Drosophila) (218974_at), score: -0.59 SPARCsecreted protein, acidic, cysteine-rich (osteonectin) (212667_at), score: 0.4 SPATS2spermatogenesis associated, serine-rich 2 (218324_s_at), score: -0.65 SPHK1sphingosine kinase 1 (219257_s_at), score: 0.41 SPON2spondin 2, extracellular matrix protein (218638_s_at), score: 0.43 SPSB1splA/ryanodine receptor domain and SOCS box containing 1 (219677_at), score: -0.57 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.56 STARD3StAR-related lipid transfer (START) domain containing 3 (202991_at), score: 0.4 STMN2stathmin-like 2 (203000_at), score: 0.41 SUSD5sushi domain containing 5 (214954_at), score: -0.67 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.59 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.61 TBC1D4TBC1 domain family, member 4 (203386_at), score: 0.47 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.65 TBL1XR1transducin (beta)-like 1 X-linked receptor 1 (221428_s_at), score: -0.62 TBPL1TBP-like 1 (208398_s_at), score: -0.63 TBX5T-box 5 (207155_at), score: 0.54 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.73 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: -0.73 TESK1testis-specific kinase 1 (204106_at), score: 0.47 TGFAtransforming growth factor, alpha (205016_at), score: 0.6 THNSL2threonine synthase-like 2 (S. cerevisiae) (219044_at), score: 0.4 THY1Thy-1 cell surface antigen (208850_s_at), score: 0.41 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.76 TLN2talin 2 (212703_at), score: 0.53 TMEM158transmembrane protein 158 (213338_at), score: 0.41 TMEM204transmembrane protein 204 (219315_s_at), score: 0.59 TMEM214transmembrane protein 214 (217899_at), score: 0.4 TMEM47transmembrane protein 47 (209656_s_at), score: 0.55 TMEM51transmembrane protein 51 (218815_s_at), score: -0.63 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.64 TMOD1tropomodulin 1 (203661_s_at), score: -0.66 TOB1transducer of ERBB2, 1 (202704_at), score: 0.55 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: -0.6 TRIM16tripartite motif-containing 16 (204341_at), score: -0.62 TRIM2tripartite motif-containing 2 (202342_s_at), score: 0.39 TSC22D1TSC22 domain family, member 1 (215111_s_at), score: -0.59 TSPAN13tetraspanin 13 (217979_at), score: 0.8 TSPYL5TSPY-like 5 (213122_at), score: 0.5 TXNRD1thioredoxin reductase 1 (201266_at), score: -0.68 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: 0.75 UBE2D4ubiquitin-conjugating enzyme E2D 4 (putative) (218837_s_at), score: 0.43 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: -0.66 UBL3ubiquitin-like 3 (201535_at), score: 0.78 UGCGUDP-glucose ceramide glucosyltransferase (204881_s_at), score: -0.66 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.71 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.76 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.77 UROSuroporphyrinogen III synthase (203031_s_at), score: 0.4 USP18ubiquitin specific peptidase 18 (219211_at), score: -0.66 UXS1UDP-glucuronate decarboxylase 1 (219675_s_at), score: 0.42 VKORC1vitamin K epoxide reductase complex, subunit 1 (217949_s_at), score: 0.54 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.59 WNT5Bwingless-type MMTV integration site family, member 5B (221029_s_at), score: 0.66 XYLT1xylosyltransferase I (213725_x_at), score: 0.61 YIPF2Yip1 domain family, member 2 (65086_at), score: 0.59 ZBTB7Azinc finger and BTB domain containing 7A (219186_at), score: 0.41 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: 0.61 ZNF281zinc finger protein 281 (218401_s_at), score: 0.42 ZNF415zinc finger protein 415 (205514_at), score: 0.44 ZNF536zinc finger protein 536 (206403_at), score: 0.58 ZNF654zinc finger protein 654 (219239_s_at), score: 0.42 ZNF804Azinc finger protein 804A (215767_at), score: -0.58
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
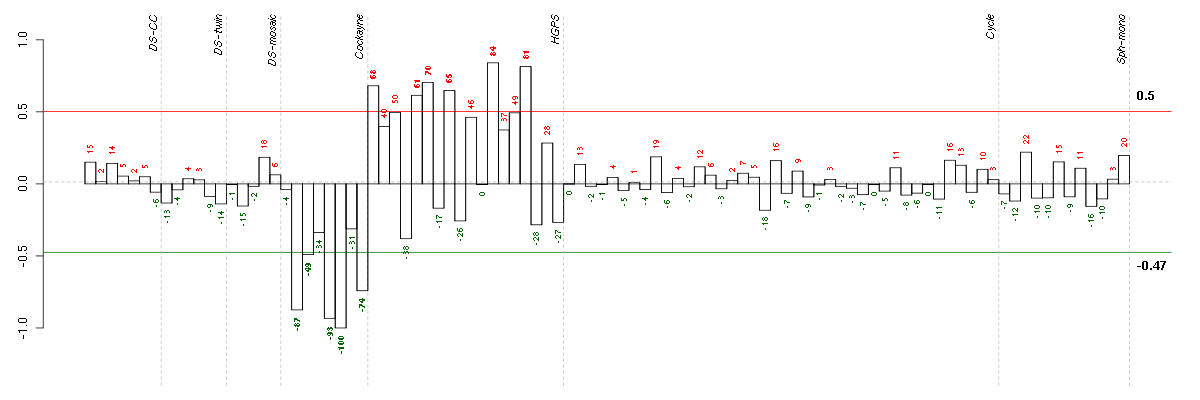
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |