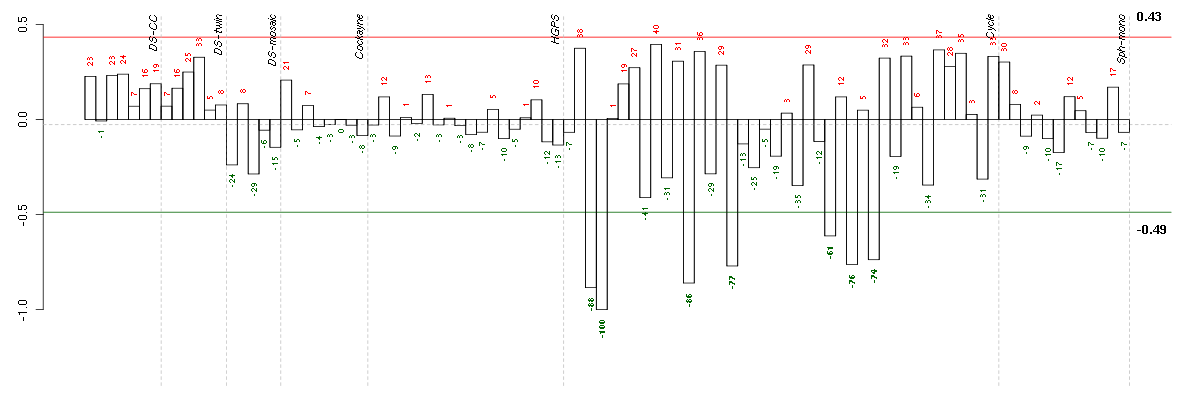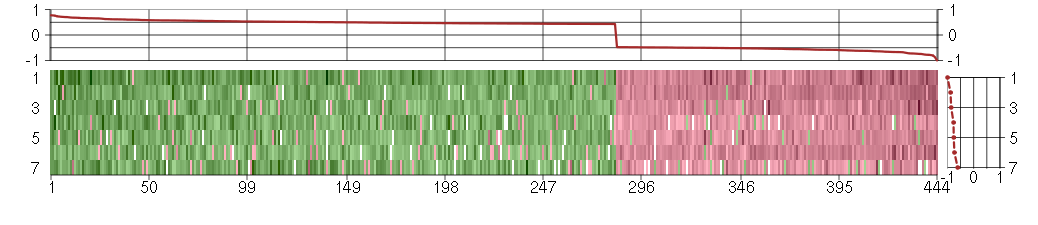

Under-expression is coded with green,
over-expression with red color.

signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.57 ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (208161_s_at), score: 0.59 ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: -0.67 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: 0.57 ACTN3actinin, alpha 3 (206891_at), score: 0.6 ACVR2Aactivin A receptor, type IIA (205327_s_at), score: 0.51 ADAadenosine deaminase (204639_at), score: 0.44 ADORA1adenosine A1 receptor (216220_s_at), score: 0.56 AFF2AF4/FMR2 family, member 2 (206105_at), score: 0.47 AGPAT31-acylglycerol-3-phosphate O-acyltransferase 3 (219723_x_at), score: -0.48 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: 0.47 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.64 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.53 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: 0.45 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.45 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: 0.44 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.54 ANKRD26ankyrin repeat domain 26 (205706_s_at), score: -0.53 ANXA8L2annexin A8-like 2 (203074_at), score: 0.5 AOC2amine oxidase, copper containing 2 (retina-specific) (207064_s_at), score: 0.52 AP1G2adaptor-related protein complex 1, gamma 2 subunit (201613_s_at), score: 0.44 APH1Aanterior pharynx defective 1 homolog A (C. elegans) (218389_s_at), score: -0.49 APOC1apolipoprotein C-I (213553_x_at), score: 0.5 APOLD1apolipoprotein L domain containing 1 (221031_s_at), score: -0.51 APPamyloid beta (A4) precursor protein (200602_at), score: 0.47 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: -0.55 ARHGDIBRho GDP dissociation inhibitor (GDI) beta (201288_at), score: -0.67 ARHGEF4Rho guanine nucleotide exchange factor (GEF) 4 (205109_s_at), score: 0.47 ARL8BADP-ribosylation factor-like 8B (217852_s_at), score: 0.65 ASB13ankyrin repeat and SOCS box-containing 13 (218862_at), score: 0.43 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: -0.5 ATF5activating transcription factor 5 (204999_s_at), score: -0.5 ATF7activating transcription factor 7 (206684_s_at), score: 0.43 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.45 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: 0.46 ATMataxia telangiectasia mutated (210858_x_at), score: 0.57 ATP6V1G2ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G2 (214762_at), score: 0.53 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.5 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.66 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.58 BCL11BB-cell CLL/lymphoma 11B (zinc finger protein) (219528_s_at), score: 0.44 BCL2L1BCL2-like 1 (215037_s_at), score: -0.59 BCL6B-cell CLL/lymphoma 6 (203140_at), score: 0.46 BEX1brain expressed, X-linked 1 (218332_at), score: -0.49 BNC1basonuclin 1 (206581_at), score: 0.45 BTNL3butyrophilin-like 3 (217207_s_at), score: 0.44 C10orf72chromosome 10 open reading frame 72 (213381_at), score: 0.46 C14orf94chromosome 14 open reading frame 94 (218383_at), score: -0.62 C16orf53chromosome 16 open reading frame 53 (218300_at), score: -0.5 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.48 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.62 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.5 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.72 C7orf44chromosome 7 open reading frame 44 (209445_x_at), score: -0.48 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.76 C9orf33chromosome 9 open reading frame 33 (217130_at), score: 0.46 C9orf91chromosome 9 open reading frame 91 (221865_at), score: 0.44 CA4carbonic anhydrase IV (206209_s_at), score: 0.5 CALB2calbindin 2 (205428_s_at), score: 0.66 CAMK1calcium/calmodulin-dependent protein kinase I (204392_at), score: -0.51 CAPN5calpain 5 (205166_at), score: -0.61 CCBP2chemokine binding protein 2 (206887_at), score: 0.43 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.47 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.55 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 0.46 CCNJcyclin J (219470_x_at), score: 0.47 CD53CD53 molecule (203416_at), score: 0.51 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.48 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: -0.54 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.58 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: -0.6 CHEK2CHK2 checkpoint homolog (S. pombe) (210416_s_at), score: -0.55 CHST1carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 (205567_at), score: 0.52 CICcapicua homolog (Drosophila) (212784_at), score: -0.53 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.58 CLCN6chloride channel 6 (203950_s_at), score: 0.51 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: -0.54 CNNM1cyclin M1 (220166_at), score: 0.44 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.58 CNR2cannabinoid receptor 2 (macrophage) (206586_at), score: 0.48 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.53 COL16A1collagen, type XVI, alpha 1 (204345_at), score: 0.59 COL4A3collagen, type IV, alpha 3 (Goodpasture antigen) (222073_at), score: 0.46 COQ10Bcoenzyme Q10 homolog B (S. cerevisiae) (219397_at), score: 0.43 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: 0.45 CPA4carboxypeptidase A4 (205832_at), score: 0.46 CPNE3copine III (202118_s_at), score: -0.66 CREG1cellular repressor of E1A-stimulated genes 1 (201200_at), score: 0.49 CRIP2cysteine-rich protein 2 (208978_at), score: -0.58 CRISPLD2cysteine-rich secretory protein LCCL domain containing 2 (221541_at), score: 0.48 CScitrate synthase (208660_at), score: -0.5 CTNScystinosis, nephropathic (36566_at), score: 0.57 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: 0.45 CXorf21chromosome X open reading frame 21 (220252_x_at), score: 0.6 CXorf56chromosome X open reading frame 56 (219805_at), score: 0.45 CYP24A1cytochrome P450, family 24, subfamily A, polypeptide 1 (206504_at), score: 0.44 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.73 CYTH4cytohesin 4 (219183_s_at), score: 0.54 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (219179_at), score: 0.45 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: -0.64 DENND3DENN/MADD domain containing 3 (212974_at), score: -0.73 DHCR77-dehydrocholesterol reductase (201790_s_at), score: 0.44 DHRS11dehydrogenase/reductase (SDR family) member 11 (218756_s_at), score: -0.51 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: 0.57 DLEU1deleted in lymphocytic leukemia 1 (non-protein coding) (205677_s_at), score: -0.48 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.61 DMC1DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) (208382_s_at), score: -0.62 DNAJC2DnaJ (Hsp40) homolog, subfamily C, member 2 (213097_s_at), score: 0.62 DNM2dynamin 2 (202253_s_at), score: -0.58 DOK4docking protein 4 (209691_s_at), score: -0.51 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.57 DUS2Ldihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae) (47105_at), score: 0.5 ECE1endothelin converting enzyme 1 (201750_s_at), score: -0.5 ECM1extracellular matrix protein 1 (209365_s_at), score: -0.5 EDARectodysplasin A receptor (220048_at), score: 0.45 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: -0.56 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: -0.5 EMCNendomucin (219436_s_at), score: -0.74 ENGendoglin (201809_s_at), score: -0.48 EPAGearly lymphoid activation protein (217050_at), score: 0.55 EPHA4EPH receptor A4 (206114_at), score: 0.44 EPHB6EPH receptor B6 (204718_at), score: 0.46 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: 0.6 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (207347_at), score: -0.61 EVLEnah/Vasp-like (217838_s_at), score: -0.55 EXOGendo/exonuclease (5'-3'), endonuclease G-like (205521_at), score: -0.56 F11coagulation factor XI (206610_s_at), score: 0.64 F2RL1coagulation factor II (thrombin) receptor-like 1 (213506_at), score: -0.48 F8A1coagulation factor VIII-associated (intronic transcript) 1 (203274_at), score: -0.48 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: 0.51 FAM118Afamily with sequence similarity 118, member A (219629_at), score: 0.45 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.58 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: 0.54 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.66 FBXO2F-box protein 2 (219305_x_at), score: 0.47 FGF12fibroblast growth factor 12 (207501_s_at), score: 0.54 FGL1fibrinogen-like 1 (205305_at), score: 0.6 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.5 FLCNfolliculin (215645_at), score: 0.45 FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.71 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: -0.78 FNDC4fibronectin type III domain containing 4 (218843_at), score: -0.51 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: -0.48 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.54 FRG1FSHD region gene 1 (204145_at), score: 0.46 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.43 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: 0.7 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.77 GADD45Bgrowth arrest and DNA-damage-inducible, beta (207574_s_at), score: 0.54 GAGE6G antigen 6 (208155_x_at), score: 0.44 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.68 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.57 GDF9growth differentiation factor 9 (221314_at), score: 0.49 GMPRguanosine monophosphate reductase (204187_at), score: -0.49 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: 0.59 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: 0.46 GOLGA8Agolgi autoantigen, golgin subfamily a, 8A (208798_x_at), score: 0.55 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: 0.7 GOLGA9Pgolgi autoantigen, golgin subfamily a, 9 pseudogene (213737_x_at), score: 0.43 GPD2glycerol-3-phosphate dehydrogenase 2 (mitochondrial) (210007_s_at), score: -0.58 GPR137BG protein-coupled receptor 137B (204137_at), score: 0.43 GPR144G protein-coupled receptor 144 (216289_at), score: 0.76 GPR32G protein-coupled receptor 32 (221469_at), score: 0.5 GPR56G protein-coupled receptor 56 (212070_at), score: -0.68 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.54 GRIA3glutamate receptor, ionotrophic, AMPA 3 (206730_at), score: 0.51 GRLF1glucocorticoid receptor DNA binding factor 1 (202044_at), score: -0.5 GSTA1glutathione S-transferase alpha 1 (215766_at), score: 0.58 GSTM5glutathione S-transferase mu 5 (205752_s_at), score: 0.58 GSTT2glutathione S-transferase theta 2 (205439_at), score: -0.5 GTF2H3general transcription factor IIH, polypeptide 3, 34kDa (222104_x_at), score: 0.54 HAB1B1 for mucin (215778_x_at), score: 0.69 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.57 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: 0.71 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: 0.52 HERC3hect domain and RLD 3 (206183_s_at), score: -0.52 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.5 HIP1Rhuntingtin interacting protein 1 related (209558_s_at), score: 0.49 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.67 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.52 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.72 HOXD4homeobox D4 (205522_at), score: 0.5 HS3ST2heparan sulfate (glucosamine) 3-O-sulfotransferase 2 (219697_at), score: 0.49 HSD17B2hydroxysteroid (17-beta) dehydrogenase 2 (204818_at), score: 0.44 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: 0.53 HSPB3heat shock 27kDa protein 3 (206375_s_at), score: 0.46 HTR1F5-hydroxytryptamine (serotonin) receptor 1F (221458_at), score: 0.46 HYIhydroxypyruvate isomerase homolog (E. coli) (221435_x_at), score: -0.49 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: 0.61 IFI30interferon, gamma-inducible protein 30 (201422_at), score: -0.58 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.56 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: 0.47 IGLL3immunoglobulin lambda-like polypeptide 3 (215946_x_at), score: 0.45 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (207901_at), score: -0.5 IL1R1interleukin 1 receptor, type I (202948_at), score: 0.51 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: 0.51 IL24interleukin 24 (206569_at), score: 0.49 IL9Rinterleukin 9 receptor (217212_s_at), score: 0.62 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.66 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.44 INTS1integrator complex subunit 1 (212212_s_at), score: -0.6 ITGA6integrin, alpha 6 (215177_s_at), score: -0.59 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.55 JAG2jagged 2 (32137_at), score: 0.65 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: -0.57 JMJD7jumonji domain containing 7 (60528_at), score: 0.44 JUPjunction plakoglobin (201015_s_at), score: -0.64 KBTBD11kelch repeat and BTB (POZ) domain containing 11 (204301_at), score: 0.44 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: 0.47 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.44 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: 0.51 KCTD2potassium channel tetramerisation domain containing 2 (212564_at), score: -0.48 KIAA0892KIAA0892 (212505_s_at), score: -0.54 KIAA1539KIAA1539 (207765_s_at), score: -0.49 KIAA1659KIAA1659 protein (215674_at), score: 0.53 KIF20Akinesin family member 20A (218755_at), score: -0.49 KIF26Bkinesin family member 26B (220002_at), score: 0.58 KLF12Kruppel-like factor 12 (208467_at), score: -0.5 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: 0.66 LAT2linker for activation of T cells family, member 2 (211768_at), score: 0.53 LEPREL2leprecan-like 2 (204854_at), score: -0.72 LIG1ligase I, DNA, ATP-dependent (202726_at), score: -0.48 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.58 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.51 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: -0.61 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: 0.44 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.65 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.48 LOC391132similar to hCG2041276 (216177_at), score: 0.56 LOC81691exonuclease NEF-sp (208107_s_at), score: -0.64 LPCAT3lysophosphatidylcholine acyltransferase 3 (202793_at), score: 0.46 LPCAT4lysophosphatidylcholine acyltransferase 4 (40472_at), score: -0.53 LPHN1latrophilin 1 (203488_at), score: 0.49 LPIN2lipin 2 (202459_s_at), score: -0.48 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.57 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.62 LXNlatexin (218729_at), score: -0.48 MACROD1MACRO domain containing 1 (219188_s_at), score: 0.55 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: 0.53 MAGEB2melanoma antigen family B, 2 (206218_at), score: 0.6 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.51 MAP2microtubule-associated protein 2 (210015_s_at), score: 0.45 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: -0.61 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.71 MAPK10mitogen-activated protein kinase 10 (204813_at), score: 0.55 MAPK6mitogen-activated protein kinase 6 (207121_s_at), score: 0.44 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: -0.52 MDKmidkine (neurite growth-promoting factor 2) (209035_at), score: 0.45 MED15mediator complex subunit 15 (222175_s_at), score: -0.52 MEG3maternally expressed 3 (non-protein coding) (210794_s_at), score: 0.65 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.73 MFSD11major facilitator superfamily domain containing 11 (221192_x_at), score: 0.51 MMEmembrane metallo-endopeptidase (203434_s_at), score: 0.45 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.47 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.52 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: 0.57 MPHOSPH10M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) (212885_at), score: 0.44 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.58 MPP2membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) (213270_at), score: 0.48 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.62 MTMR11myotubularin related protein 11 (205076_s_at), score: -0.62 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.43 MYH15myosin, heavy chain 15 (215331_at), score: 0.44 MYL10myosin, light chain 10, regulatory (221659_s_at), score: 0.5 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.43 MYO15Amyosin XVA (220288_at), score: 0.52 NAB2NGFI-A binding protein 2 (EGR1 binding protein 2) (216017_s_at), score: -0.62 NACA2nascent polypeptide-associated complex alpha subunit 2 (222224_at), score: 0.44 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: 0.49 NARG1LNMDA receptor regulated 1-like (219378_at), score: 0.52 NCAM1neural cell adhesion molecule 1 (212843_at), score: 0.52 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: -0.5 NCLNnicalin homolog (zebrafish) (222206_s_at), score: -0.52 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.51 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: 0.48 NCRNA00092non-protein coding RNA 92 (215861_at), score: 0.53 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: 0.56 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.67 NF1neurofibromin 1 (211094_s_at), score: -0.67 NMT2N-myristoyltransferase 2 (205005_s_at), score: -0.48 NOL10nucleolar protein 10 (218591_s_at), score: 0.48 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.49 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: -0.57 NUAK1NUAK family, SNF1-like kinase, 1 (204589_at), score: 0.45 NXPH3neurexophilin 3 (221991_at), score: 0.49 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.62 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: -0.5 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: -0.56 OLFML2Bolfactomedin-like 2B (213125_at), score: -0.87 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.77 OPRL1opiate receptor-like 1 (206564_at), score: 0.47 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.77 PAPSS23'-phosphoadenosine 5'-phosphosulfate synthase 2 (203059_s_at), score: -0.51 PARP6poly (ADP-ribose) polymerase family, member 6 (219639_x_at), score: 0.58 PARVBparvin, beta (37965_at), score: -0.54 PBX3pre-B-cell leukemia homeobox 3 (204082_at), score: -0.49 PCDH24protocadherin 24 (220186_s_at), score: 0.57 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.49 PCNXpecanex homolog (Drosophila) (213159_at), score: -0.53 PCSK5proprotein convertase subtilisin/kexin type 5 (205559_s_at), score: 0.57 PDE4Dphosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) (204491_at), score: 0.43 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.74 PDGFDplatelet derived growth factor D (219304_s_at), score: 0.5 PGCprogastricsin (pepsinogen C) (205261_at), score: 0.5 PHF15PHD finger protein 15 (212660_at), score: -0.49 PHF7PHD finger protein 7 (215622_x_at), score: 0.53 PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: 0.54 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.51 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.55 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.48 PLAUplasminogen activator, urokinase (211668_s_at), score: -0.58 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: -0.49 PMLpromyelocytic leukemia (206503_x_at), score: -0.55 PMS2PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (209805_at), score: 0.45 PNMAL1PNMA-like 1 (218824_at), score: -0.63 POLEpolymerase (DNA directed), epsilon (216026_s_at), score: -0.49 POLR2J4polymerase (RNA) II (DNA directed) polypeptide J4, pseudogene (217610_at), score: 0.47 POU4F1POU class 4 homeobox 1 (206940_s_at), score: 0.52 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: 0.69 PRLHprolactin releasing hormone (221443_x_at), score: 0.45 PRRX1paired related homeobox 1 (205991_s_at), score: -0.65 PTDSS1phosphatidylserine synthase 1 (201433_s_at), score: -0.48 PTGESprostaglandin E synthase (210367_s_at), score: 0.45 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: -0.49 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.67 PXNpaxillin (211823_s_at), score: -0.79 QPRTquinolinate phosphoribosyltransferase (204044_at), score: 0.46 RAPGEF2Rap guanine nucleotide exchange factor (GEF) 2 (203097_s_at), score: 0.44 RASA4RAS p21 protein activator 4 (208534_s_at), score: -0.52 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: 0.66 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -1 REG3Aregenerating islet-derived 3 alpha (205815_at), score: 0.51 RNF121ring finger protein 121 (219021_at), score: 0.47 RNF220ring finger protein 220 (219988_s_at), score: -0.49 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.54 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.57 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: -0.53 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.47 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: -0.58 RREB1ras responsive element binding protein 1 (203704_s_at), score: 0.56 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: 0.55 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: -0.5 RUNX1runt-related transcription factor 1 (209360_s_at), score: 0.48 SAA4serum amyloid A4, constitutive (207096_at), score: 0.61 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.53 SC4MOLsterol-C4-methyl oxidase-like (209146_at), score: 0.51 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.54 SCDstearoyl-CoA desaturase (delta-9-desaturase) (200832_s_at), score: 0.54 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: -0.8 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.55 SEMA4Gsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4G (219194_at), score: 0.45 SEPT8septin 8 (209000_s_at), score: -0.55 SFTPBsurfactant protein B (214354_x_at), score: 0.5 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.57 SHPKsedoheptulokinase (219713_at), score: 0.45 SHROOM2shroom family member 2 (204967_at), score: 0.56 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: 0.51 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: 0.51 SLC11A2solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 (203124_s_at), score: 0.5 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: 0.58 SLC22A17solute carrier family 22, member 17 (218675_at), score: 0.5 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.62 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.73 SLC2A1solute carrier family 2 (facilitated glucose transporter), member 1 (201250_s_at), score: 0.46 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: 0.61 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: 0.44 SLC2A4RGSLC2A4 regulator (218494_s_at), score: -0.51 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: 0.53 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: -0.55 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.52 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: -0.48 SNTG1syntrophin, gamma 1 (220405_at), score: -0.5 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.51 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: 0.68 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.51 SPATA1spermatogenesis associated 1 (221057_at), score: 0.52 SPATA2Lspermatogenesis associated 2-like (214965_at), score: 0.46 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: 0.44 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: -0.5 SRFserum response factor (c-fos serum response element-binding transcription factor) (202401_s_at), score: 0.46 ST3GAL2ST3 beta-galactoside alpha-2,3-sialyltransferase 2 (217650_x_at), score: 0.43 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.46 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.54 STARD3StAR-related lipid transfer (START) domain containing 3 (202991_at), score: -0.48 STK16serine/threonine kinase 16 (209622_at), score: 0.46 STX3syntaxin 3 (209238_at), score: 0.49 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.56 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: 0.47 SYT1synaptotagmin I (203999_at), score: -0.54 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.46 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: 0.48 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.61 TDGthymine-DNA glycosylase (203743_s_at), score: 0.47 TFDP1transcription factor Dp-1 (204147_s_at), score: -0.56 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.78 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: 0.54 TGM4transglutaminase 4 (prostate) (217566_s_at), score: 0.61 THRAthyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) (35846_at), score: 0.53 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: 0.5 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: 0.53 TMEM39Btransmembrane protein 39B (218770_s_at), score: -0.53 TMEM48transmembrane protein 48 (218073_s_at), score: -0.5 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.62 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: 0.48 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.51 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: -0.66 TOMM34translocase of outer mitochondrial membrane 34 (201870_at), score: -0.49 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: 0.66 TRIM3tripartite motif-containing 3 (204911_s_at), score: -0.52 TRIM34tripartite motif-containing 34 (221044_s_at), score: -0.53 TRY6trypsinogen C (215395_x_at), score: -0.55 TULP2tubby like protein 2 (206733_at), score: 0.62 TXNthioredoxin (216609_at), score: 0.48 TYMPthymidine phosphorylase (217497_at), score: 0.47 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: -0.52 URB2URB2 ribosome biogenesis 2 homolog (S. cerevisiae) (205284_at), score: 0.5 UTF1undifferentiated embryonic cell transcription factor 1 (208275_x_at), score: 0.48 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.53 VDRvitamin D (1,25- dihydroxyvitamin D3) receptor (204255_s_at), score: 0.5 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.69 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.47 WDR3WD repeat domain 3 (218882_s_at), score: 0.47 WDR42AWD repeat domain 42A (202249_s_at), score: -0.52 WDR60WD repeat domain 60 (219251_s_at), score: 0.49 WDR62WD repeat domain 62 (215218_s_at), score: -0.52 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: 0.44 WNT5Awingless-type MMTV integration site family, member 5A (213425_at), score: 0.6 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: 0.45 WSB1WD repeat and SOCS box-containing 1 (201294_s_at), score: 0.51 YAF2YY1 associated factor 2 (206238_s_at), score: 0.47 YIPF1Yip1 domain family, member 1 (214733_s_at), score: 0.5 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.49 ZAKsterile alpha motif and leucine zipper containing kinase AZK (218833_at), score: 0.45 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.43 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.63 ZNF281zinc finger protein 281 (218401_s_at), score: 0.52 ZNF492zinc finger protein 492 (215532_x_at), score: 0.44 ZNF510zinc finger protein 510 (206053_at), score: -0.64 ZNF587zinc finger protein 587 (219981_x_at), score: 0.44 ZNF652zinc finger protein 652 (205594_at), score: 0.68 ZNF767zinc finger family member 767 (219627_at), score: 0.44 ZNF783zinc finger family member 783 (221876_at), score: 0.54
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |