
Under-expression is coded with green,
over-expression with red color.



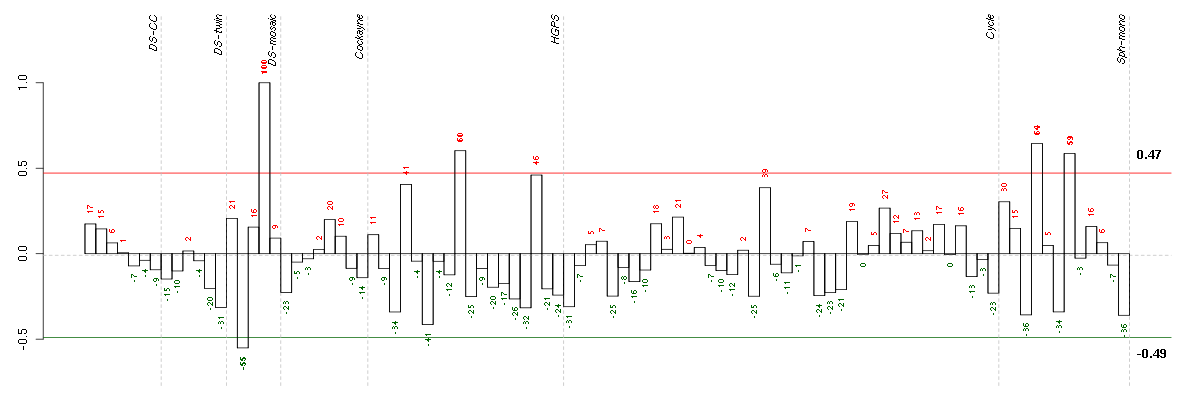
CHKAcholine kinase alpha (204233_s_at), score: 0.77 HDAC9histone deacetylase 9 (205659_at), score: 0.83 KCNAB1potassium voltage-gated channel, shaker-related subfamily, beta member 1 (210078_s_at), score: 0.89 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: 1 SLC46A3solute carrier family 46, member 3 (214719_at), score: 0.96
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |