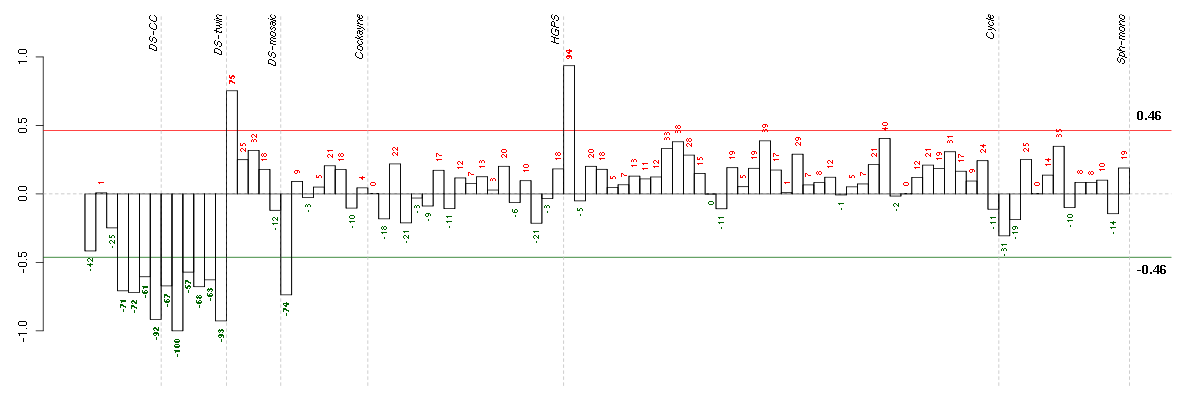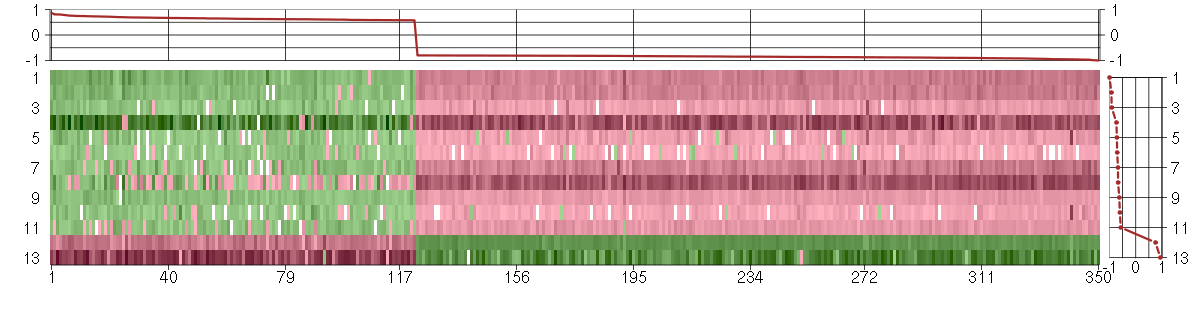

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell motion
Any process involved in the controlled movement of a cell.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
cell surface receptor linked signal transduction
Any series of molecular signals initiated by the binding of an extracellular ligand to a receptor on the surface of the target cell.
G-protein coupled receptor protein signaling pathway
The series of molecular signals generated as a consequence of a G-protein coupled receptor binding to its physiological ligand.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell migration
The orderly movement of cells from one site to another, often during the development of a multicellular organism or multicellular structure.
antigen processing and presentation
The process by which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
locomotion
Self-propelled movement of a cell or organism from one location to another.
antigen processing and presentation of peptide antigen
The process by which an antigen-presenting cell expresses peptide antigen in association with an MHC protein complex on its cell surface, including proteolysis and transport steps for the peptide antigen both prior to and following assembly with the MHC protein complex. The peptide antigen is typically, but not always, processed from an endogenous or exogenous protein.
cell motility
Any process involved in the controlled movement of a cell that results in translocation of the cell from one place to another.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
keratinocyte migration
The directed movement of keratinocytes, epidermal cells which synthesize keratin, from one site to another.
localization of cell
Any process by which a cell is transported to, and/or maintained in, a specific location.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cell motility
Any process involved in the controlled movement of a cell that results in translocation of the cell from one place to another.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
cell motion
Any process involved in the controlled movement of a cell.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05330 | 9.104e-03 | 0.8125 | 6 | 21 | Allograft rejection |
| 05332 | 9.104e-03 | 0.8125 | 6 | 21 | Graft-versus-host disease |
| 05320 | 1.138e-02 | 0.8512 | 6 | 22 | Autoimmune thyroid disease |
ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: -0.82 ABI3BPABI family, member 3 (NESH) binding protein (220518_at), score: -0.82 ACRV1acrosomal vesicle protein 1 (207969_x_at), score: -0.8 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: 0.65 ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: -0.84 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.88 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: -0.91 ADAMTSL4ADAMTS-like 4 (220578_at), score: -0.81 ADCY2adenylate cyclase 2 (brain) (217687_at), score: -0.84 ADCY6adenylate cyclase 6 (209195_s_at), score: 0.58 ADD2adducin 2 (beta) (206807_s_at), score: -0.82 ADIPOR2adiponectin receptor 2 (201346_at), score: 0.64 AGBL2ATP/GTP binding protein-like 2 (220390_at), score: -0.84 AGMATagmatine ureohydrolase (agmatinase) (219792_at), score: -0.96 AJAP1adherens junctions associated protein 1 (206460_at), score: 0.69 AKAP6A kinase (PRKA) anchor protein 6 (205359_at), score: -0.81 ALDOBaldolase B, fructose-bisphosphate (214423_x_at), score: -0.93 ANXA2P3annexin A2 pseudogene 3 (211241_at), score: -0.91 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (204650_s_at), score: -0.81 APOMapolipoprotein M (205682_x_at), score: -0.99 ARL17P1ADP-ribosylation factor-like 17 pseudogene 1 (210718_s_at), score: -0.8 ART1ADP-ribosyltransferase 1 (207919_at), score: -0.8 ATP8A1ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 (213106_at), score: -0.81 ATRNL1attractin-like 1 (213744_at), score: -0.84 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (221234_s_at), score: -0.8 BCANbrevican (91920_at), score: -0.82 BCL7AB-cell CLL/lymphoma 7A (203795_s_at), score: 0.63 BCORL1BCL6 co-repressor-like 1 (219444_at), score: -0.9 BEND5BEN domain containing 5 (219670_at), score: -0.97 BMP10bone morphogenetic protein 10 (208292_at), score: -0.84 BMPR2bone morphogenetic protein receptor, type II (serine/threonine kinase) (210214_s_at), score: 0.64 BRD3bromodomain containing 3 (203825_at), score: -0.87 BRS3bombesin-like receptor 3 (207369_at), score: -0.93 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.9 C11orf1chromosome 11 open reading frame 1 (218925_s_at), score: -0.84 C11orf21chromosome 11 open reading frame 21 (220560_at), score: -0.86 C14orf93chromosome 14 open reading frame 93 (219009_at), score: -0.81 C17orf71chromosome 17 open reading frame 71 (218514_at), score: 0.6 C19orf28chromosome 19 open reading frame 28 (220178_at), score: 0.68 C1orf159chromosome 1 open reading frame 159 (219337_at), score: -0.84 C1orf61chromosome 1 open reading frame 61 (205103_at), score: -0.96 C1orf69chromosome 1 open reading frame 69 (215490_at), score: -0.81 C2orf54chromosome 2 open reading frame 54 (220149_at), score: -0.89 C8orf17chromosome 8 open reading frame 17 (208266_at), score: -0.87 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.84 C9orf33chromosome 9 open reading frame 33 (217130_at), score: -0.94 CA1carbonic anhydrase I (205949_at), score: -0.83 CA4carbonic anhydrase IV (206209_s_at), score: -0.9 CAPN9calpain 9 (210641_at), score: -0.83 CARTPTCART prepropeptide (206339_at), score: -0.83 CASP4caspase 4, apoptosis-related cysteine peptidase (209310_s_at), score: 0.69 CASZ1castor zinc finger 1 (220015_at), score: -0.91 CCR9chemokine (C-C motif) receptor 9 (207445_s_at), score: -0.89 CD302CD302 molecule (203799_at), score: 0.62 CD3EAPCD3e molecule, epsilon associated protein (205264_at), score: 0.59 CD40CD40 molecule, TNF receptor superfamily member 5 (35150_at), score: -0.9 CD46CD46 molecule, complement regulatory protein (211574_s_at), score: 0.58 CD53CD53 molecule (203416_at), score: -0.81 CDC42SE1CDC42 small effector 1 (218157_x_at), score: 0.71 CDH18cadherin 18, type 2 (206280_at), score: -0.87 CDH8cadherin 8, type 2 (217574_at), score: -0.8 CDYLchromodomain protein, Y-like (203100_s_at), score: 0.61 CEACAM1carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) (211883_x_at), score: -0.91 CEACAM5carcinoembryonic antigen-related cell adhesion molecule 5 (201884_at), score: -0.89 CEACAM6carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) (211657_at), score: -0.87 CEP152centrosomal protein 152kDa (215170_s_at), score: -0.88 CEP27centrosomal protein 27kDa (220071_x_at), score: -0.87 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: 0.59 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.92 CLDN18claudin 18 (214135_at), score: -0.88 CLEC1AC-type lectin domain family 1, member A (219761_at), score: -0.81 CLEC4MC-type lectin domain family 4, member M (207995_s_at), score: -0.84 CLEC7AC-type lectin domain family 7, member A (221698_s_at), score: -0.9 CNTLNcentlein, centrosomal protein (220095_at), score: -0.83 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.88 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.81 CR1complement component (3b/4b) receptor 1 (Knops blood group) (217484_at), score: -0.9 CREG1cellular repressor of E1A-stimulated genes 1 (201200_at), score: 0.69 CRYGDcrystallin, gamma D (207532_at), score: -0.88 CSN2casein beta (207951_at), score: -0.8 CST3cystatin C (201360_at), score: 0.59 CTAGE1cutaneous T-cell lymphoma-associated antigen 1 (220957_at), score: -0.81 CTSEcathepsin E (205927_s_at), score: -0.88 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: -0.91 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (216809_at), score: -0.83 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: -0.92 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: -0.86 CYP2E1cytochrome P450, family 2, subfamily E, polypeptide 1 (1431_at), score: -0.86 DAOD-amino-acid oxidase (206878_at), score: -0.91 DAZ1deleted in azoospermia 1 (216922_x_at), score: -0.81 DCP1ADCP1 decapping enzyme homolog A (S. cerevisiae) (218508_at), score: 0.58 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: 0.61 DESdesmin (216947_at), score: -0.85 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.63 DNASE1L3deoxyribonuclease I-like 3 (205554_s_at), score: -0.81 DNASE2deoxyribonuclease II, lysosomal (214992_s_at), score: 0.68 DNMBPdynamin binding protein (212838_at), score: 0.6 DNMT3BDNA (cytosine-5-)-methyltransferase 3 beta (220668_s_at), score: -0.83 DNTTIP2deoxynucleotidyltransferase, terminal, interacting protein 2 (202776_at), score: 0.7 DOCK9dedicator of cytokinesis 9 (212538_at), score: 0.62 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: 0.8 DRD5dopamine receptor D5 (208486_at), score: -0.87 DTX4deltex homolog 4 (Drosophila) (212611_at), score: -0.81 EDARectodysplasin A receptor (220048_at), score: -0.85 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: 0.59 ELNelastin (212670_at), score: -0.85 EPHB3EPH receptor B3 (1438_at), score: -0.86 EPORerythropoietin receptor (396_f_at), score: -0.82 EXD3exonuclease 3'-5' domain containing 3 (220838_at), score: -0.87 F10coagulation factor X (205620_at), score: 0.78 FABP7fatty acid binding protein 7, brain (205030_at), score: -0.86 FAM38Bfamily with sequence similarity 38, member B (219602_s_at), score: -0.81 FAM75A3family with sequence similarity 75, member A3 (215935_at), score: -0.86 FASTKD5FAST kinase domains 5 (219016_at), score: 0.58 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: 0.67 FLGfilaggrin (215704_at), score: -0.95 FMO4flavin containing monooxygenase 4 (206263_at), score: -0.94 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.58 GABRR1gamma-aminobutyric acid (GABA) receptor, rho 1 (206525_at), score: -0.85 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (208217_at), score: -0.86 GALgalanin prepropeptide (214240_at), score: 0.59 GAP43growth associated protein 43 (204471_at), score: -0.82 GK2glycerol kinase 2 (215430_at), score: -0.81 GLTSCR1glioma tumor suppressor candidate region gene 1 (219445_at), score: -0.85 GNALguanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type (206355_at), score: -0.9 GNAO1guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O (204762_s_at), score: -0.84 GNG4guanine nucleotide binding protein (G protein), gamma 4 (205184_at), score: -0.89 GNRHRgonadotropin-releasing hormone receptor (216341_s_at), score: -0.9 GPD1Lglycerol-3-phosphate dehydrogenase 1-like (212510_at), score: 0.62 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: 0.7 GPR12G protein-coupled receptor 12 (214558_at), score: -0.84 GPR183G protein-coupled receptor 183 (205419_at), score: 0.65 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: 0.64 GSTT1glutathione S-transferase theta 1 (203815_at), score: 0.69 HABP2hyaluronan binding protein 2 (206010_at), score: -0.85 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.89 HAVCR1hepatitis A virus cellular receptor 1 (207052_at), score: -0.85 HBEGFheparin-binding EGF-like growth factor (203821_at), score: 0.68 hCG_1732469hCG1732469 (220823_at), score: -0.81 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.58 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: 0.59 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: 0.67 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.85 HLA-Amajor histocompatibility complex, class I, A (213932_x_at), score: 0.64 HLA-Amajor histocompatibility complex, class I, A (217436_x_at), score: 0.66 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: 0.68 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.74 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: 0.73 HMGB3L1high-mobility group box 3-like 1 (216548_x_at), score: -0.8 HOXD1homeobox D1 (205975_s_at), score: -0.81 HOXD11homeobox D11 (214604_at), score: -0.99 HOXD4homeobox D4 (205522_at), score: -0.95 HPCAL1hippocalcin-like 1 (205462_s_at), score: 0.66 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: -0.82 HTR1E5-hydroxytryptamine (serotonin) receptor 1E (207404_s_at), score: -0.81 IFITM1interferon induced transmembrane protein 1 (9-27) (214022_s_at), score: 0.63 IFNA7interferon, alpha 7 (208259_x_at), score: -0.81 IFNGinterferon, gamma (210354_at), score: -0.82 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: -0.8 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.66 IL6interleukin 6 (interferon, beta 2) (205207_at), score: 0.71 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.8 ITIH2inter-alpha (globulin) inhibitor H2 (204987_at), score: -0.96 ITKIL2-inducible T-cell kinase (211339_s_at), score: -0.88 KCNIP1Kv channel interacting protein 1 (221307_at), score: -0.86 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: -0.81 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (219287_at), score: -0.96 KIAA0317KIAA0317 (202128_at), score: 0.6 KIDINS220kinase D-interacting substrate, 220kDa (212162_at), score: 0.6 KIF5Akinesin family member 5A (205318_at), score: -0.86 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.83 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.95 KLHL21kelch-like 21 (Drosophila) (203068_at), score: 0.58 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.62 KRT2keratin 2 (207908_at), score: -0.83 L3MBTLl(3)mbt-like (Drosophila) (210306_at), score: -0.81 LAT2linker for activation of T cells family, member 2 (211768_at), score: -0.85 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: -0.8 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.62 LHX3LIM homeobox 3 (221670_s_at), score: -0.85 LILRB4leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4 (210152_at), score: -0.83 LITAFlipopolysaccharide-induced TNF factor (200706_s_at), score: 0.67 LMBRD1LMBR1 domain containing 1 (218191_s_at), score: 0.58 LOC100130233similar to NPM1 protein (216387_x_at), score: -0.85 LOC100130703similar to hCG2042168 (207596_at), score: -0.82 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: -0.85 LOC150759hypothetical protein LOC150759 (213703_at), score: -0.8 LOC389906similar to Serine/threonine-protein kinase PRKX (Protein kinase PKX1) (59433_at), score: -0.81 LOC730227hypothetical protein LOC730227 (215756_at), score: -0.95 LONRF3LON peptidase N-terminal domain and ring finger 3 (220009_at), score: -0.89 LYNv-yes-1 Yamaguchi sarcoma viral related oncogene homolog (202625_at), score: 0.75 MADCAM1mucosal vascular addressin cell adhesion molecule 1 (208037_s_at), score: -0.82 MAGI2membrane associated guanylate kinase, WW and PDZ domain containing 2 (209737_at), score: 0.59 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.76 MAN2B1mannosidase, alpha, class 2B, member 1 (209166_s_at), score: 0.61 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.66 MBmyoglobin (204179_at), score: -1 MCTP2multiple C2 domains, transmembrane 2 (220603_s_at), score: -0.82 MEIS1Meis homeobox 1 (204069_at), score: 0.65 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: 0.74 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.88 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.82 MGC13053hypothetical MGC13053 (211775_x_at), score: -0.81 MIA2melanoma inhibitory activity 2 (221177_at), score: -0.81 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (203936_s_at), score: -0.87 MPP3membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) (206186_at), score: -0.83 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.87 MTSS1metastasis suppressor 1 (203037_s_at), score: 0.63 MUC3Amucin 3A, cell surface associated (217117_x_at), score: -0.88 MUSKmuscle, skeletal, receptor tyrosine kinase (207633_s_at), score: -0.84 MYO5Cmyosin VC (218966_at), score: -0.85 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.82 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.87 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: 0.59 NFIBnuclear factor I/B (209290_s_at), score: -0.86 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.87 NID1nidogen 1 (202008_s_at), score: 0.69 NINninein (GSK3B interacting protein) (219285_s_at), score: -0.84 NINJ1ninjurin 1 (203045_at), score: 0.73 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: -0.87 NOVA1neuro-oncological ventral antigen 1 (205794_s_at), score: -0.95 NPnucleoside phosphorylase (201695_s_at), score: 0.61 NPC1Niemann-Pick disease, type C1 (202679_at), score: 0.63 NPY2Rneuropeptide Y receptor Y2 (210730_s_at), score: -0.85 NR3C1nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (201866_s_at), score: 0.62 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: 0.58 NRN1neuritin 1 (218625_at), score: -0.88 NTNG1netrin G1 (206713_at), score: -0.83 OCLMoculomedin (208274_at), score: -0.81 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.62 OR10C1olfactory receptor, family 10, subfamily C, member 1 (221339_at), score: -0.94 OR1D2olfactory receptor, family 1, subfamily D, member 2 (221464_at), score: -0.86 OR7E24olfactory receptor, family 7, subfamily E, member 24 (215463_at), score: -0.88 OVOL2ovo-like 2 (Drosophila) (211778_s_at), score: -0.86 PCDH7protocadherin 7 (205534_at), score: 0.62 PCYT1Aphosphate cytidylyltransferase 1, choline, alpha (204210_s_at), score: 0.61 PECAM1platelet/endothelial cell adhesion molecule (208982_at), score: -0.93 PEG3paternally expressed 3 (209242_at), score: -0.97 PER1period homolog 1 (Drosophila) (36829_at), score: -0.92 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.89 PGLYRP4peptidoglycan recognition protein 4 (220944_at), score: -0.81 PIP5K1Bphosphatidylinositol-4-phosphate 5-kinase, type I, beta (217477_at), score: -0.82 PKP2plakophilin 2 (207717_s_at), score: 0.61 PLUNCpalate, lung and nasal epithelium associated (220542_s_at), score: -0.88 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: 0.73 PMP2peripheral myelin protein 2 (206826_at), score: -0.81 PMS2L3postmeiotic segregation increased 2-like 3 (214473_x_at), score: 0.61 POLR1Cpolymerase (RNA) I polypeptide C, 30kDa (207515_s_at), score: 0.7 PON1paraoxonase 1 (206344_at), score: -0.81 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.92 PPARDperoxisome proliferator-activated receptor delta (37152_at), score: 0.81 PPP4R4protein phosphatase 4, regulatory subunit 4 (220672_at), score: -0.81 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: -0.9 PRKCBprotein kinase C, beta (209685_s_at), score: -0.88 PROL1proline rich, lacrimal 1 (208004_at), score: -0.92 PROX1prospero homeobox 1 (207401_at), score: -0.91 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: 0.79 PYHIN1pyrin and HIN domain family, member 1 (216748_at), score: -0.86 RAB26RAB26, member RAS oncogene family (50965_at), score: -0.88 RAD9ARAD9 homolog A (S. pombe) (204828_at), score: -0.82 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: -0.93 RAMP1receptor (G protein-coupled) activity modifying protein 1 (204916_at), score: 0.67 RCAN2regulator of calcineurin 2 (203498_at), score: 0.61 RCL1RNA terminal phosphate cyclase-like 1 (218544_s_at), score: 0.65 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.81 RETret proto-oncogene (205879_x_at), score: -0.81 RFPL3ret finger protein-like 3 (207936_x_at), score: -0.87 RFX3regulatory factor X, 3 (influences HLA class II expression) (207234_at), score: -0.82 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: -0.87 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.93 RPS14ribosomal protein S14 (208646_at), score: -0.8 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.72 RRADRas-related associated with diabetes (204802_at), score: 0.66 RRHretinal pigment epithelium-derived rhodopsin homolog (208314_at), score: -0.95 RUNX1runt-related transcription factor 1 (209360_s_at), score: 0.66 S100A12S100 calcium binding protein A12 (205863_at), score: -0.89 S100A14S100 calcium binding protein A14 (218677_at), score: -0.87 SASH3SAM and SH3 domain containing 3 (204923_at), score: -0.83 SCN10Asodium channel, voltage-gated, type X, alpha subunit (208578_at), score: -0.81 SCYL2SCY1-like 2 (S. cerevisiae) (221220_s_at), score: 0.58 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: 0.65 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: -0.89 SERPINB13serpin peptidase inhibitor, clade B (ovalbumin), member 13 (211362_s_at), score: -0.8 SERPINB8serpin peptidase inhibitor, clade B (ovalbumin), member 8 (206034_at), score: 0.63 SETXsenataxin (201964_at), score: 0.63 SH3BGRLSH3 domain binding glutamic acid-rich protein like (201311_s_at), score: 0.58 SH3TC2SH3 domain and tetratricopeptide repeats 2 (219710_at), score: -0.81 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.66 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.89 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (207095_at), score: -0.86 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: -0.83 SLC15A1solute carrier family 15 (oligopeptide transporter), member 1 (211349_at), score: -0.86 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (209681_at), score: 0.67 SLC1A7solute carrier family 1 (glutamate transporter), member 7 (210923_at), score: -0.87 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.86 SLC4A5solute carrier family 4, sodium bicarbonate cotransporter, member 5 (204296_at), score: -0.91 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: -0.8 SP140SP140 nuclear body protein (207777_s_at), score: -0.8 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.85 SPEGSPEG complex locus (205265_s_at), score: -0.81 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: 0.6 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: 0.65 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.63 SRFserum response factor (c-fos serum response element-binding transcription factor) (202401_s_at), score: 0.62 STARD13StAR-related lipid transfer (START) domain containing 13 (213103_at), score: 0.58 STK38Lserine/threonine kinase 38 like (212572_at), score: 0.62 STOMstomatin (201060_x_at), score: 0.68 SVEP1sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (213247_at), score: 0.74 TAC3tachykinin 3 (219992_at), score: -0.83 TACR1tachykinin receptor 1 (208048_at), score: -0.85 TAS2R16taste receptor, type 2, member 16 (221444_at), score: -0.9 TEX2testis expressed 2 (218099_at), score: 0.66 TLR1toll-like receptor 1 (210176_at), score: -0.8 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: 0.76 TMC7transmembrane channel-like 7 (220021_at), score: -0.8 TMCO1transmembrane and coiled-coil domains 1 (210768_x_at), score: 0.58 TMEM144transmembrane protein 144 (219750_at), score: -0.83 TMEM19transmembrane protein 19 (219941_at), score: -0.88 TMEM206transmembrane protein 206 (218814_s_at), score: -0.81 TMEM39Atransmembrane protein 39A (218615_s_at), score: 0.66 TMEM41Btransmembrane protein 41B (212623_at), score: 0.65 TNFSF15tumor necrosis factor (ligand) superfamily, member 15 (221085_at), score: -0.82 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.62 TOR1AIP1torsin A interacting protein 1 (216100_s_at), score: 0.62 TP53BP2tumor protein p53 binding protein, 2 (203120_at), score: 0.75 TP63tumor protein p63 (209863_s_at), score: -0.92 TPP1tripeptidyl peptidase I (200742_s_at), score: 0.72 TPST1tyrosylprotein sulfotransferase 1 (204140_at), score: 0.64 TPST2tyrosylprotein sulfotransferase 2 (204079_at), score: 0.59 TRA@T cell receptor alpha locus (216540_at), score: -0.82 TRAF3TNF receptor-associated factor 3 (221571_at), score: 0.63 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: 0.67 TRIM10tripartite motif-containing 10 (56748_at), score: -0.88 TRIM45tripartite motif-containing 45 (219923_at), score: -0.95 TRPM3transient receptor potential cation channel, subfamily M, member 3 (220463_at), score: -0.85 TSPAN14tetraspanin 14 (221002_s_at), score: -0.81 TULP2tubby like protein 2 (206733_at), score: -0.81 UBE3Bubiquitin protein ligase E3B (212403_at), score: 0.62 UTP3UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) (209486_at), score: 0.63 WDR3WD repeat domain 3 (218882_s_at), score: 0.69 WDR78WD repeat domain 78 (220769_s_at), score: -0.83 YRDCyrdC domain containing (E. coli) (218647_s_at), score: 0.72 ZIC1Zic family member 1 (odd-paired homolog, Drosophila) (206373_at), score: -0.84 ZKSCAN3zinc finger with KRAB and SCAN domains 3 (211773_s_at), score: -0.86 ZNF286Azinc finger protein 286A (220250_at), score: -0.85 ZNF287zinc finger protein 287 (216453_at), score: -0.92 ZNF35zinc finger protein 35 (206096_at), score: 0.74 ZNF362zinc finger protein 362 (214915_at), score: -0.82 ZNF672zinc finger protein 672 (218068_s_at), score: 0.68 ZPBPzona pellucida binding protein (207021_at), score: -0.82
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |