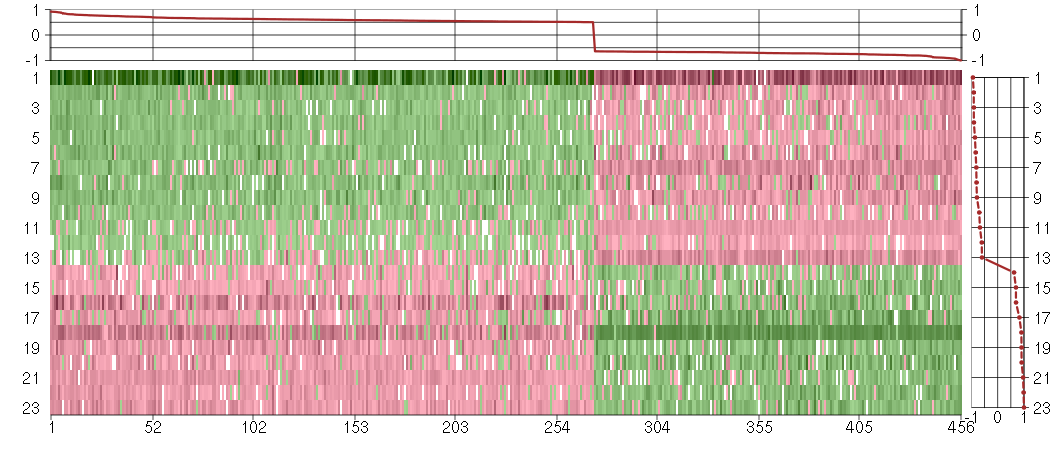

Under-expression is coded with green,
over-expression with red color.

signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.72 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.76 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.62 ABLIM3actin binding LIM protein family, member 3 (205730_s_at), score: 0.63 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.64 ACTN3actinin, alpha 3 (206891_at), score: -0.74 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.71 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.7 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.54 AESamino-terminal enhancer of split (217729_s_at), score: 0.55 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.56 AGRNagrin (212285_s_at), score: 0.55 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -1 ANK2ankyrin 2, neuronal (202920_at), score: 0.53 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: 0.55 ANXA2P1annexin A2 pseudogene 1 (210876_at), score: -0.68 ANXA8L2annexin A8-like 2 (203074_at), score: -0.77 APOC4apolipoprotein C-IV (206738_at), score: -0.74 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.66 ARMC9armadillo repeat containing 9 (219637_at), score: 0.6 ARSAarylsulfatase A (204443_at), score: 0.76 ART1ADP-ribosyltransferase 1 (207919_at), score: -0.68 ATF5activating transcription factor 5 (204999_s_at), score: 0.55 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.69 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: 0.6 B9D1B9 protein domain 1 (210534_s_at), score: 0.6 BAIAP3BAI1-associated protein 3 (216356_x_at), score: -0.69 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.65 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.8 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.72 BCANbrevican (91920_at), score: -0.67 BCL2L1BCL2-like 1 (215037_s_at), score: 0.58 BDH23-hydroxybutyrate dehydrogenase, type 2 (218285_s_at), score: 0.62 BDKRB2bradykinin receptor B2 (205870_at), score: 0.55 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: 0.51 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.6 BTG2BTG family, member 2 (201236_s_at), score: 0.6 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.69 C10orf119chromosome 10 open reading frame 119 (217905_at), score: -0.65 C10orf26chromosome 10 open reading frame 26 (202808_at), score: 0.55 C14orf159chromosome 14 open reading frame 159 (218298_s_at), score: 0.53 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.7 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.63 C1orf89chromosome 1 open reading frame 89 (220963_s_at), score: -0.67 C22orf30chromosome 22 open reading frame 30 (216555_at), score: -0.71 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.68 C8orf60chromosome 8 open reading frame 60 (220712_at), score: -0.68 C9orf167chromosome 9 open reading frame 167 (219620_x_at), score: 0.51 CALB1calbindin 1, 28kDa (205626_s_at), score: -0.74 CALB2calbindin 2 (205428_s_at), score: -0.98 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.76 CASP2caspase 2, apoptosis-related cysteine peptidase (209811_at), score: -0.66 CASZ1castor zinc finger 1 (220015_at), score: -0.69 CCDC101coiled-coil domain containing 101 (48117_at), score: -0.75 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.73 CD40CD40 molecule, TNF receptor superfamily member 5 (35150_at), score: -0.66 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.79 CDAcytidine deaminase (205627_at), score: 0.63 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.64 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.57 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (218062_x_at), score: 0.66 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: -0.8 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (220446_s_at), score: -0.67 CICcapicua homolog (Drosophila) (212784_at), score: 0.61 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.67 CLDN1claudin 1 (218182_s_at), score: 0.5 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.55 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.78 CNNM1cyclin M1 (220166_at), score: -0.66 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.57 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.87 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.79 CORO1Bcoronin, actin binding protein, 1B (64486_at), score: 0.64 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: -0.65 CRCPCGRP receptor component (203898_at), score: -0.75 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.74 CSNK2A2casein kinase 2, alpha prime polypeptide (203575_at), score: 0.55 CTSEcathepsin E (205927_s_at), score: -0.79 CUL7cullin 7 (36084_at), score: 0.64 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: -0.66 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.72 CYTH4cytohesin 4 (219183_s_at), score: -0.69 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: -0.67 DDR1discoidin domain receptor tyrosine kinase 1 (208779_x_at), score: 0.63 DEAF1deformed epidermal autoregulatory factor 1 (Drosophila) (209407_s_at), score: 0.6 DENND3DENN/MADD domain containing 3 (212974_at), score: 0.59 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.67 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.78 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.72 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.88 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.74 DNM2dynamin 2 (202253_s_at), score: 0.55 DOK4docking protein 4 (209691_s_at), score: 0.56 DOPEY1dopey family member 1 (40612_at), score: -0.77 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: 0.61 DSPdesmoplakin (200606_at), score: -0.69 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.8 DUS2Ldihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae) (47105_at), score: -0.72 ECE1endothelin converting enzyme 1 (201750_s_at), score: 0.61 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.72 EFHD2EF-hand domain family, member D2 (217992_s_at), score: 0.52 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.55 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.84 EPB41L5erythrocyte membrane protein band 4.1 like 5 (220977_x_at), score: -0.67 EPHA4EPH receptor A4 (206114_at), score: -0.89 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.64 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: -0.65 EPN2epsin 2 (203463_s_at), score: 0.53 ERAP2endoplasmic reticulum aminopeptidase 2 (219759_at), score: 0.65 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.67 ETV7ets variant 7 (221680_s_at), score: -0.69 EVLEnah/Vasp-like (217838_s_at), score: 0.57 EXD3exonuclease 3'-5' domain containing 3 (220838_at), score: -0.74 EXOC2exocyst complex component 2 (219349_s_at), score: 0.58 F11coagulation factor XI (206610_s_at), score: -0.64 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.67 FAM176Bfamily with sequence similarity 176, member B (220134_x_at), score: 0.51 FAM62Afamily with sequence similarity 62 (C2 domain containing), member A (208858_s_at), score: 0.54 FAM65Afamily with sequence similarity 65, member A (45749_at), score: 0.58 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: -0.72 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: 0.58 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.75 FBXO17F-box protein 17 (220233_at), score: 0.61 FBXO2F-box protein 2 (219305_x_at), score: -0.71 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.8 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.73 FDXRferredoxin reductase (207813_s_at), score: 0.51 FETUBfetuin B (214417_s_at), score: -0.82 FGF12fibroblast growth factor 12 (207501_s_at), score: -0.66 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.52 FGL1fibrinogen-like 1 (205305_at), score: -0.7 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.67 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.61 FKSG2apoptosis inhibitor (208588_at), score: -0.67 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.66 FNBP1formin binding protein 1 (212288_at), score: 0.52 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.66 FOXF1forkhead box F1 (205935_at), score: 0.67 FOXK2forkhead box K2 (203064_s_at), score: 0.82 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: -0.74 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (208217_at), score: -0.66 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.72 GDF9growth differentiation factor 9 (221314_at), score: -0.72 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.67 GM2AGM2 ganglioside activator (212737_at), score: 0.63 GMPRguanosine monophosphate reductase (204187_at), score: 0.79 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: -0.65 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.71 GPD1Lglycerol-3-phosphate dehydrogenase 1-like (212510_at), score: 0.58 GRAMD1BGRAM domain containing 1B (212906_at), score: -0.67 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.54 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.53 GTPBP1GTP binding protein 1 (219357_at), score: 0.64 HAB1B1 for mucin (215778_x_at), score: -0.96 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.9 HAO2hydroxyacid oxidase 2 (long chain) (220801_s_at), score: -0.72 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: -0.72 HDAC5histone deacetylase 5 (202455_at), score: 0.54 HECTD3HECT domain containing 3 (218632_at), score: 0.64 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.67 HHLA3HERV-H LTR-associating 3 (220387_s_at), score: 0.56 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.68 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.72 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.73 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.75 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.6 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.58 HOXC5homeobox C5 (206739_at), score: -0.65 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.81 HSF1heat shock transcription factor 1 (202344_at), score: 0.72 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.62 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.53 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.91 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.75 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.74 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: -0.72 IL9Rinterleukin 9 receptor (217212_s_at), score: -0.91 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.82 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.61 IRF1interferon regulatory factor 1 (202531_at), score: 0.58 IRS4insulin receptor substrate 4 (207403_at), score: -0.68 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: 0.58 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.65 JMJD7jumonji domain containing 7 (60528_at), score: -0.65 JUNDjun D proto-oncogene (203751_x_at), score: 0.76 JUPjunction plakoglobin (201015_s_at), score: 0.89 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.93 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: -0.66 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.56 KIAA0317KIAA0317 (202128_at), score: 0.54 KIAA0556KIAA0556 (213185_at), score: 0.51 KIAA0913KIAA0913 (212359_s_at), score: 0.64 KIAA1305KIAA1305 (220911_s_at), score: 0.9 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.8 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.64 KRT24keratin 24 (220267_at), score: -0.7 KSR1kinase suppressor of ras 1 (213769_at), score: -0.72 LEPREL2leprecan-like 2 (204854_at), score: 0.74 LILRB4leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4 (210152_at), score: -0.64 LIMS2LIM and senescent cell antigen-like domains 2 (220765_s_at), score: 0.57 LITAFlipopolysaccharide-induced TNF factor (200706_s_at), score: 0.61 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.77 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.73 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.66 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.54 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.65 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.53 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: 0.62 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.58 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.78 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.55 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.85 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.78 MAP2microtubule-associated protein 2 (210015_s_at), score: -0.69 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.52 MAP2K3mitogen-activated protein kinase kinase 3 (207667_s_at), score: 0.58 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: 0.57 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.69 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.54 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.52 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.74 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.59 MGST2microsomal glutathione S-transferase 2 (204168_at), score: 0.64 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.54 MKL1megakaryoblastic leukemia (translocation) 1 (212748_at), score: 0.52 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.67 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.67 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.65 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.67 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.59 MPP3membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) (206186_at), score: -0.64 MRPS2mitochondrial ribosomal protein S2 (218001_at), score: 0.56 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.53 MUC13mucin 13, cell surface associated (218687_s_at), score: -0.67 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.79 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.67 MYH15myosin, heavy chain 15 (215331_at), score: -0.66 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.92 MYO15Amyosin XVA (220288_at), score: -0.64 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.71 NCALDneurocalcin delta (211685_s_at), score: -0.66 NCDNneurochondrin (209556_at), score: 0.58 NCK2NCK adaptor protein 2 (203315_at), score: 0.59 NCKAP1NCK-associated protein 1 (207738_s_at), score: -0.8 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.71 NEFMneurofilament, medium polypeptide (205113_at), score: -0.89 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.65 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.67 NINJ1ninjurin 1 (203045_at), score: 0.77 NMIN-myc (and STAT) interactor (203964_at), score: 0.56 NOL10nucleolar protein 10 (218591_s_at), score: -0.76 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: 0.5 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: -0.66 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.81 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.61 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.58 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.67 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: -0.69 NXPH3neurexophilin 3 (221991_at), score: -0.78 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: 0.54 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: 0.52 OPRL1opiate receptor-like 1 (206564_at), score: -0.67 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: -0.8 OVOL2ovo-like 2 (Drosophila) (211778_s_at), score: -0.71 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.53 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: -0.64 PARVBparvin, beta (37965_at), score: 0.72 PBX3pre-B-cell leukemia homeobox 3 (204082_at), score: 0.52 PCDH9protocadherin 9 (219737_s_at), score: -0.78 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.71 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.57 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.68 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.65 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.77 PDE7Bphosphodiesterase 7B (220343_at), score: -0.76 PDK1pyruvate dehydrogenase kinase, isozyme 1 (206686_at), score: -0.65 PDK2pyruvate dehydrogenase kinase, isozyme 2 (202590_s_at), score: 0.52 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.67 PHF7PHD finger protein 7 (215622_x_at), score: -0.88 PHLDA3pleckstrin homology-like domain, family A, member 3 (218634_at), score: 0.6 PI4K2Aphosphatidylinositol 4-kinase type 2 alpha (209345_s_at), score: 0.63 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.53 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (207105_s_at), score: 0.51 PINK1PTEN induced putative kinase 1 (209018_s_at), score: 0.57 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.84 PKN1protein kinase N1 (202161_at), score: 0.78 PKP2plakophilin 2 (207717_s_at), score: 0.53 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: 0.52 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.72 PLD3phospholipase D family, member 3 (201050_at), score: 0.56 PLEKHM2pleckstrin homology domain containing, family M (with RUN domain) member 2 (212146_at), score: 0.56 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.52 PNMAL1PNMA-like 1 (218824_at), score: 0.66 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.61 PNRC1proline-rich nuclear receptor coactivator 1 (209034_at), score: 0.52 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.63 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: 0.54 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.58 PORP450 (cytochrome) oxidoreductase (208928_at), score: 0.68 PRKCBprotein kinase C, beta (209685_s_at), score: -0.66 PRKCDprotein kinase C, delta (202545_at), score: 0.71 PRLHprolactin releasing hormone (221443_x_at), score: -0.74 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.65 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.57 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.78 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.68 PXNpaxillin (211823_s_at), score: 0.66 PYGBphosphorylase, glycogen; brain (201481_s_at), score: 0.61 PYGLphosphorylase, glycogen, liver (202990_at), score: 0.57 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.61 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.54 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.66 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.63 RABL4RAB, member of RAS oncogene family-like 4 (205037_at), score: 0.6 RANBP3RAN binding protein 3 (210120_s_at), score: 0.51 RBM15BRNA binding motif protein 15B (202689_at), score: 0.55 RCAN2regulator of calcineurin 2 (203498_at), score: 0.51 REEP1receptor accessory protein 1 (204364_s_at), score: -0.71 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.75 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.62 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.56 RGS2regulator of G-protein signaling 2, 24kDa (202388_at), score: 0.55 RHBDF1rhomboid 5 homolog 1 (Drosophila) (218686_s_at), score: 0.59 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.56 RLN1relaxin 1 (211753_s_at), score: -0.72 RNF11ring finger protein 11 (208924_at), score: -0.65 RNF130ring finger protein 130 (217865_at), score: 0.67 RNF220ring finger protein 220 (219988_s_at), score: 0.65 RNF24ring finger protein 24 (204669_s_at), score: 0.66 RNF40ring finger protein 40 (206845_s_at), score: 0.54 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.63 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.74 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.79 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.69 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.78 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.76 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.7 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.9 S100A14S100 calcium binding protein A14 (218677_at), score: -0.71 SAA4serum amyloid A4, constitutive (207096_at), score: -0.7 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.81 SBF1SET binding factor 1 (39835_at), score: 0.51 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.65 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.91 SCAMP5secretory carrier membrane protein 5 (212699_at), score: -0.69 SCAPSREBF chaperone (212329_at), score: 0.54 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.65 SDC3syndecan 3 (202898_at), score: 0.63 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.62 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.54 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.74 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.51 SF1splicing factor 1 (208313_s_at), score: 0.51 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.59 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.7 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.68 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.81 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.8 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.88 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.72 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.67 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: 0.51 SLC38A10solute carrier family 38, member 10 (212890_at), score: 0.55 SLC41A3solute carrier family 41, member 3 (219175_s_at), score: 0.52 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: 0.54 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.72 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.89 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.63 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.67 SMAD3SMAD family member 3 (218284_at), score: 0.59 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.72 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.5 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.71 SMYD5SMYD family member 5 (209516_at), score: 0.54 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.74 SNIP1Smad nuclear interacting protein 1 (219409_at), score: 0.54 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.68 SPAG8sperm associated antigen 8 (206816_s_at), score: -0.66 SPARCL1SPARC-like 1 (hevin) (200795_at), score: -0.74 SPATA1spermatogenesis associated 1 (221057_at), score: -0.67 SPATA20spermatogenesis associated 20 (218164_at), score: 0.74 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.63 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: 0.52 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.6 SSX3synovial sarcoma, X breakpoint 3 (215881_x_at), score: -0.65 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.62 STIM1stromal interaction molecule 1 (202764_at), score: 0.64 STOMstomatin (201060_x_at), score: 0.59 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.88 SUOXsulfite oxidase (204067_at), score: 0.58 SYNPOsynaptopodin (202796_at), score: 0.63 SYT1synaptotagmin I (203999_at), score: 0.59 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.74 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.74 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: -0.66 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.77 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.73 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.66 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.73 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.52 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.74 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.76 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.69 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.53 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.78 TLR3toll-like receptor 3 (206271_at), score: 0.62 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.75 TMEM115transmembrane protein 115 (216267_s_at), score: 0.52 TMEM35transmembrane protein 35 (219685_at), score: 0.52 TMEM40transmembrane protein 40 (219503_s_at), score: -0.65 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.78 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.71 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.53 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.54 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.62 TNS3tensin 3 (217853_at), score: 0.51 TOB2transducer of ERBB2, 2 (221496_s_at), score: 0.58 TP53tumor protein p53 (201746_at), score: 0.62 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: 0.6 TPP1tripeptidyl peptidase I (200742_s_at), score: 0.52 TRA@T cell receptor alpha locus (216540_at), score: -0.84 TRIM21tripartite motif-containing 21 (204804_at), score: 0.64 TRIM45tripartite motif-containing 45 (219923_at), score: -0.72 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.73 TSKUtsukushin (218245_at), score: 0.75 TSPAN2tetraspanin 2 (214606_at), score: -0.67 TSSC4tumor suppressing subtransferable candidate 4 (218612_s_at), score: 0.51 TTC15tetratricopeptide repeat domain 15 (203122_at), score: 0.55 TXLNAtaxilin alpha (212300_at), score: 0.58 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.67 UBAC1UBA domain containing 1 (202151_s_at), score: 0.6 UBAP1ubiquitin associated protein 1 (221490_at), score: 0.52 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.71 UBXN1UBX domain protein 1 (201871_s_at), score: 0.51 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.57 VAT1vesicle amine transport protein 1 homolog (T. californica) (208626_s_at), score: 0.57 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.74 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.91 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.82 WBP2WW domain binding protein 2 (209117_at), score: 0.71 WDR4WD repeat domain 4 (221632_s_at), score: -0.76 WDR42AWD repeat domain 42A (202249_s_at), score: 0.69 WDR6WD repeat domain 6 (217734_s_at), score: 0.8 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.72 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: -0.67 WSB1WD repeat and SOCS box-containing 1 (201294_s_at), score: -0.66 ZAKsterile alpha motif and leucine zipper containing kinase AZK (218833_at), score: -0.66 ZDHHC11zinc finger, DHHC-type containing 11 (221646_s_at), score: -0.66 ZDHHC4zinc finger, DHHC-type containing 4 (220261_s_at), score: 0.61 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.63 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.61 ZFP106zinc finger protein 106 homolog (mouse) (217781_s_at), score: 0.52 ZNF282zinc finger protein 282 (212892_at), score: 0.6 ZNF580zinc finger protein 580 (220748_s_at), score: 0.56 ZPBPzona pellucida binding protein (207021_at), score: -0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |