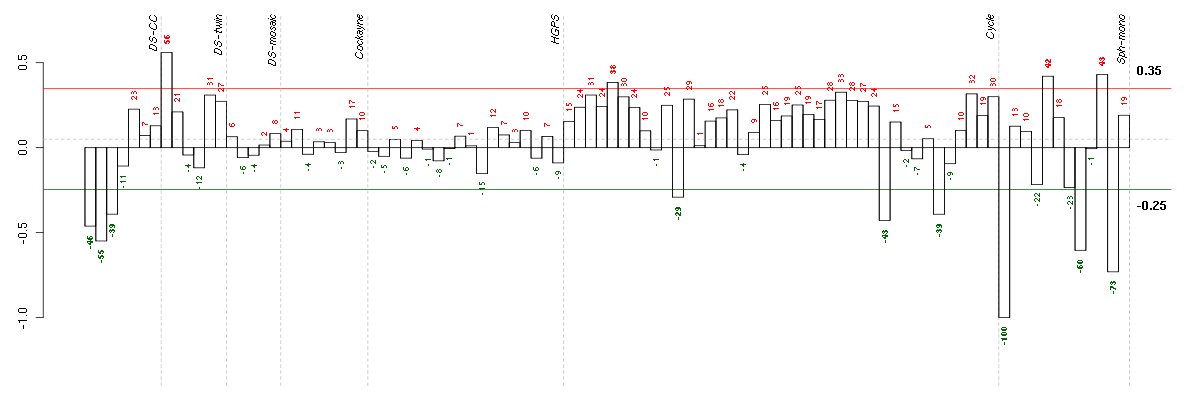

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
positive regulation of biosynthetic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
positive regulation of cellular biosynthetic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of biosynthetic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
positive regulation of biosynthetic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
positive regulation of cellular biosynthetic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
positive regulation of cellular biosynthetic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of signal transduction
Any process that modulates the frequency, rate or extent of signal transduction.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
positive regulation of cellular biosynthetic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
G-protein-coupled receptor binding
Interacting selectively with a G-protein-coupled receptor.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively with any nucleic acid.
DNA binding
Interacting selectively with DNA (deoxyribonucleic acid).
transcription factor activity
The function of binding to a specific DNA sequence in order to modulate transcription. The transcription factor may or may not also interact selectively with a protein or macromolecular complex.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
cytokine activity
Functions to control the survival, growth, differentiation and effector function of tissues and cells.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
chemokine activity
The function of a family of chemotactic pro-inflammatory activation-inducible cytokines acting primarily upon hemopoietic cells in immunoregulatory processes; all chemokines possess a number of conserved cysteine residues involved in intramolecular disulfide bond formation.
transcription regulator activity
Plays a role in regulating transcription; may bind a promoter or enhancer DNA sequence or interact with a DNA-binding transcription factor.
chemokine receptor binding
Interacting selectively with any chemokine receptor.
all
This term is the most general term possible
transcription factor activity
The function of binding to a specific DNA sequence in order to modulate transcription. The transcription factor may or may not also interact selectively with a protein or macromolecular complex.
chemokine activity
The function of a family of chemotactic pro-inflammatory activation-inducible cytokines acting primarily upon hemopoietic cells in immunoregulatory processes; all chemokines possess a number of conserved cysteine residues involved in intramolecular disulfide bond formation.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 4.170e-05 | 4.877 | 19 | 115 | Cytokine-cytokine receptor interaction |
| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-17-5p/20/93.mr/106/519.d | 1.474e-02 | 31.74 | 58 | 633 |
AASSaminoadipate-semialdehyde synthase (214829_at), score: -0.43 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (203504_s_at), score: -0.62 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (213353_at), score: -0.39 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: -0.63 ACOT11acyl-CoA thioesterase 11 (216103_at), score: -0.41 ACVR2Aactivin A receptor, type IIA (205327_s_at), score: -0.54 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: -0.33 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: -0.42 ADORA2Aadenosine A2a receptor (205013_s_at), score: -0.77 ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: -0.3 AKAP8A kinase (PRKA) anchor protein 8 (203848_at), score: -0.33 ALDH1B1aldehyde dehydrogenase 1 family, member B1 (209646_x_at), score: -0.33 ALDOAP2aldolase A, fructose-bisphosphate pseudogene 2 (211617_at), score: -0.33 ALMS1Alstrom syndrome 1 (214707_x_at), score: -0.47 AMPD3adenosine monophosphate deaminase (isoform E) (207992_s_at), score: -0.63 ANGPTL4angiopoietin-like 4 (221009_s_at), score: -0.52 ANKHankylosis, progressive homolog (mouse) (220076_at), score: -0.69 ANKRD10ankyrin repeat domain 10 (218093_s_at), score: -0.64 ANKRD12ankyrin repeat domain 12 (216550_x_at), score: -0.29 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: -0.54 APODapolipoprotein D (201525_at), score: -0.93 ARAP2ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 (214102_at), score: -0.43 AREGamphiregulin (205239_at), score: -0.44 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: -0.45 ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (202548_s_at), score: -0.3 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: -0.44 ARID4BAT rich interactive domain 4B (RBP1-like) (221230_s_at), score: -0.33 ATAD2BATPase family, AAA domain containing 2B (213387_at), score: -0.49 ATF3activating transcription factor 3 (202672_s_at), score: -0.33 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: -0.52 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221484_at), score: -0.45 BACH1BTB and CNC homology 1, basic leucine zipper transcription factor 1 (204194_at), score: -0.38 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (221234_s_at), score: -0.4 BAG5BCL2-associated athanogene 5 (202984_s_at), score: -0.29 BCL9B-cell CLL/lymphoma 9 (204129_at), score: -0.51 BCORBCL6 co-repressor (219433_at), score: -0.45 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: -0.59 BMP2bone morphogenetic protein 2 (205289_at), score: -0.79 BNC1basonuclin 1 (206581_at), score: -0.32 BTAF1BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae) (209430_at), score: -0.6 C10orf110chromosome 10 open reading frame 110 (220703_at), score: -0.29 C10orf88chromosome 10 open reading frame 88 (219240_s_at), score: -0.34 C11orf57chromosome 11 open reading frame 57 (218314_s_at), score: -0.49 C12orf35chromosome 12 open reading frame 35 (218614_at), score: -0.31 C12orf49chromosome 12 open reading frame 49 (218867_s_at), score: -0.3 C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: -0.31 C17orf91chromosome 17 open reading frame 91 (214696_at), score: -0.43 C3complement component 3 (217767_at), score: -0.44 C6orf162chromosome 6 open reading frame 162 (213314_at), score: -0.33 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: -0.55 CADPS2Ca++-dependent secretion activator 2 (219572_at), score: -0.37 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.3 CCDC25coiled-coil domain containing 25 (218125_s_at), score: -0.37 CCL7chemokine (C-C motif) ligand 7 (208075_s_at), score: -0.34 CCNJcyclin J (219470_x_at), score: -0.44 CD302CD302 molecule (203799_at), score: -0.6 CDC14ACDC14 cell division cycle 14 homolog A (S. cerevisiae) (205288_at), score: -0.48 CDC42SE1CDC42 small effector 1 (218157_x_at), score: -0.55 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213183_s_at), score: -0.51 CDR1cerebellar degeneration-related protein 1, 34kDa (207276_at), score: -0.47 CGGBP1CGG triplet repeat binding protein 1 (214050_at), score: -0.57 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: -0.53 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: -0.78 CISD3CDGSH iron sulfur domain 3 (213551_x_at), score: -0.37 CLCA2chloride channel accessory 2 (206165_s_at), score: -0.31 CLEC4EC-type lectin domain family 4, member E (219859_at), score: -0.29 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: -0.33 COL15A1collagen, type XV, alpha 1 (203477_at), score: -0.51 COL21A1collagen, type XXI, alpha 1 (208096_s_at), score: -0.35 COQ10Bcoenzyme Q10 homolog B (S. cerevisiae) (219397_at), score: -0.39 CPEcarboxypeptidase E (201117_s_at), score: -0.57 CRISPLD2cysteine-rich secretory protein LCCL domain containing 2 (221541_at), score: -0.3 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.36 CTSFcathepsin F (203657_s_at), score: -0.42 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.46 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: -0.79 CXCL2chemokine (C-X-C motif) ligand 2 (209774_x_at), score: -1 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: -0.97 CXCL5chemokine (C-X-C motif) ligand 5 (214974_x_at), score: -0.62 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.5 CXCR7chemokine (C-X-C motif) receptor 7 (212977_at), score: -0.42 CYLC1cylicin, basic protein of sperm head cytoskeleton 1 (216809_at), score: -0.29 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: -0.39 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (219179_at), score: -0.46 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: -0.5 DAZ1deleted in azoospermia 1 (216922_x_at), score: -0.31 DAZ4deleted in azoospermia 4 (216351_x_at), score: -0.32 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: -0.31 DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (206752_s_at), score: -0.3 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: -0.31 DKFZp686O1327hypothetical gene supported by BC043549; BX648102 (216877_at), score: -0.56 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: -0.58 DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: -0.49 DNAH7dynein, axonemal, heavy chain 7 (214222_at), score: -0.29 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.34 DOCK4dedicator of cytokinesis 4 (205003_at), score: -0.38 DPTdermatopontin (213068_at), score: -0.35 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.37 DUSP6dual specificity phosphatase 6 (208891_at), score: -0.35 EDNRAendothelin receptor type A (204464_s_at), score: -0.36 EEF1Deukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) (214395_x_at), score: -0.46 EGR1early growth response 1 (201694_s_at), score: -0.38 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: -0.86 EGR3early growth response 3 (206115_at), score: -0.58 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: -0.38 EIF5A2eukaryotic translation initiation factor 5A2 (220198_s_at), score: -0.4 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: -0.54 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.51 EPAGearly lymphoid activation protein (217050_at), score: -0.3 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: -0.31 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: -0.35 EREGepiregulin (205767_at), score: -0.51 ETS2v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) (201328_at), score: -0.55 EVI5ecotropic viral integration site 5 (209717_at), score: -0.37 FAM134Afamily with sequence similarity 134, member A (222129_at), score: -0.35 FAM60Afamily with sequence similarity 60, member A (220147_s_at), score: -0.38 FBXW12F-box and WD repeat domain containing 12 (215600_x_at), score: -0.3 FCARFc fragment of IgA, receptor for (211305_x_at), score: -0.33 FEM1Bfem-1 homolog b (C. elegans) (212367_at), score: -0.37 FGD6FYVE, RhoGEF and PH domain containing 6 (219901_at), score: -0.46 FLJ10038hypothetical protein FLJ10038 (205510_s_at), score: -0.5 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: -0.54 FLJ23172hypothetical LOC389177 (217016_x_at), score: -0.37 FNBP4formin binding protein 4 (212232_at), score: -0.53 FOLH1folate hydrolase (prostate-specific membrane antigen) 1 (217487_x_at), score: -0.31 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: -0.67 FOXC1forkhead box C1 (213260_at), score: -0.4 FOXN3forkhead box N3 (218031_s_at), score: -0.43 FOXO1forkhead box O1 (202724_s_at), score: -0.5 FUBP1far upstream element (FUSE) binding protein 1 (212847_at), score: -0.46 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: -0.59 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: -0.41 GCH1GTP cyclohydrolase 1 (204224_s_at), score: -0.71 GCLCglutamate-cysteine ligase, catalytic subunit (202922_at), score: -0.3 GEMGTP binding protein overexpressed in skeletal muscle (204472_at), score: -0.39 GFPT2glutamine-fructose-6-phosphate transaminase 2 (205100_at), score: -0.3 GORASP1golgi reassembly stacking protein 1, 65kDa (56919_at), score: -0.41 GPM6Bglycoprotein M6B (209167_at), score: -0.64 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: -0.37 GPR137BG protein-coupled receptor 137B (204137_at), score: -0.31 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: -0.35 GRM6glutamate receptor, metabotropic 6 (208035_at), score: -0.34 H2AFJH2A histone family, member J (220936_s_at), score: -0.3 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: -0.37 HCG2P7HLA complex group 2 pseudogene 7 (216229_x_at), score: -0.32 HECAheadcase homolog (Drosophila) (218603_at), score: -0.53 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.32 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.41 HIC2hypermethylated in cancer 2 (212964_at), score: -0.35 HIST1H2AChistone cluster 1, H2ac (215071_s_at), score: -0.39 HIST2H2BEhistone cluster 2, H2be (202708_s_at), score: -0.52 HIVEP1human immunodeficiency virus type I enhancer binding protein 1 (204512_at), score: -0.43 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: -0.8 HK2hexokinase 2 (202934_at), score: -0.61 HMGCS13-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) (221750_at), score: -0.47 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (213359_at), score: -0.56 HS2ST1heparan sulfate 2-O-sulfotransferase 1 (203285_s_at), score: -0.34 IBD12Inflammatory bowel disease 12 (215373_x_at), score: -0.39 ICAM1intercellular adhesion molecule 1 (202638_s_at), score: -0.37 IER3immediate early response 3 (201631_s_at), score: -0.58 IER3IP1immediate early response 3 interacting protein 1 (211406_at), score: -0.29 IL11interleukin 11 (206924_at), score: -0.42 IL1Ainterleukin 1, alpha (210118_s_at), score: -0.35 IL1Binterleukin 1, beta (39402_at), score: -0.6 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: -0.8 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: -0.5 IL1RNinterleukin 1 receptor antagonist (212657_s_at), score: -0.67 IL24interleukin 24 (206569_at), score: -0.49 IL6interleukin 6 (interferon, beta 2) (205207_at), score: -0.43 IL8interleukin 8 (211506_s_at), score: -0.71 IMAASLC7A5 pseudogene (208118_x_at), score: -0.3 INSRinsulin receptor (213792_s_at), score: -0.72 IREB2iron-responsive element binding protein 2 (214666_x_at), score: -0.41 IRS2insulin receptor substrate 2 (209185_s_at), score: -0.33 ISL1ISL LIM homeobox 1 (206104_at), score: -0.45 ITGB5integrin, beta 5 (214021_x_at), score: -0.36 ITPKBinositol 1,4,5-trisphosphate 3-kinase B (203723_at), score: -0.57 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: -0.4 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: -0.71 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: -0.86 JMJD1Cjumonji domain containing 1C (221763_at), score: -0.3 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: -0.84 JUNBjun B proto-oncogene (201473_at), score: -0.31 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: -0.36 KIAA0247KIAA0247 (202181_at), score: -0.73 KIAA0562KIAA0562 (204075_s_at), score: -0.31 KIAA1024KIAA1024 (215081_at), score: -0.69 KIAA1128KIAA1128 (209379_s_at), score: -0.41 KIF1Bkinesin family member 1B (209234_at), score: -0.31 KLF11Kruppel-like factor 11 (218486_at), score: -0.41 KLF9Kruppel-like factor 9 (203543_s_at), score: -0.43 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: -0.47 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: -0.4 LOC100128701heterogeneous nuclear ribonucleoprotein A1-like 2 pseudogene (216497_at), score: -0.49 LOC100128836similar to heterogeneous nuclear ribonucleoprotein A1 (217353_at), score: -0.44 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: -0.51 LOC643313similar to hypothetical protein LOC284701 (211050_x_at), score: -0.39 LOC644617hypothetical LOC644617 (221235_s_at), score: -0.57 LOC647070hypothetical LOC647070 (215467_x_at), score: -0.38 LOC65998hypothetical protein LOC65998 (218641_at), score: -0.4 LOC728844hypothetical LOC728844 (222040_at), score: -0.36 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: -0.45 LPPR4plasticity related gene 1 (213496_at), score: -0.44 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: -0.7 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: -0.37 LY75lymphocyte antigen 75 (205668_at), score: -0.32 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: -0.38 MAP3K8mitogen-activated protein kinase kinase kinase 8 (205027_s_at), score: -0.45 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: -0.37 MCL1myeloid cell leukemia sequence 1 (BCL2-related) (214057_at), score: -0.44 MCM9minichromosome maintenance complex component 9 (219673_at), score: -0.31 MDN1MDN1, midasin homolog (yeast) (212693_at), score: -0.29 MEFVMediterranean fever (208262_x_at), score: -0.36 METAP2methionyl aminopeptidase 2 (202015_x_at), score: -0.47 METTL7Amethyltransferase like 7A (211424_x_at), score: -0.47 MMEmembrane metallo-endopeptidase (203434_s_at), score: -0.35 MSL2male-specific lethal 2 homolog (Drosophila) (218733_at), score: -0.64 MSX1msh homeobox 1 (205932_s_at), score: -0.42 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (220346_at), score: -0.42 MTSS1metastasis suppressor 1 (203037_s_at), score: -0.35 MXI1MAX interactor 1 (202364_at), score: -0.54 MYNNmyoneurin (218926_at), score: -0.36 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: -0.6 NAMPTnicotinamide phosphoribosyltransferase (217738_at), score: -0.65 NBEAneurobeachin (221207_s_at), score: -0.28 NET1neuroepithelial cell transforming 1 (201830_s_at), score: -0.32 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: -0.3 NIPSNAP3Bnipsnap homolog 3B (C. elegans) (221104_s_at), score: -0.29 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: -0.36 NR4A2nuclear receptor subfamily 4, group A, member 2 (216248_s_at), score: -0.55 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: -0.79 NUMBnumb homolog (Drosophila) (207545_s_at), score: -0.38 NUP214nucleoporin 214kDa (202155_s_at), score: -0.37 NUPL1nucleoporin like 1 (204435_at), score: -0.39 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.34 OR2B2olfactory receptor, family 2, subfamily B, member 2 (216408_at), score: -0.31 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.55 PAPPApregnancy-associated plasma protein A, pappalysin 1 (201981_at), score: -0.3 PARP8poly (ADP-ribose) polymerase family, member 8 (219033_at), score: -0.4 PBX2pre-B-cell leukemia homeobox 2 (202876_s_at), score: -0.53 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: -0.44 PCMTD2protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 (212406_s_at), score: -0.49 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: -0.33 PELI1pellino homolog 1 (Drosophila) (218319_at), score: -0.61 PELI2pellino homolog 2 (Drosophila) (219132_at), score: -0.74 PER2period homolog 2 (Drosophila) (205251_at), score: -0.51 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (202464_s_at), score: -0.58 PHF2PHD finger protein 2 (212726_at), score: -0.35 PHF21APHD finger protein 21A (203278_s_at), score: -0.36 PHF8PHD finger protein 8 (212916_at), score: -0.4 PHLPPPH domain and leucine rich repeat protein phosphatase (212719_at), score: -0.36 PIAS2protein inhibitor of activated STAT, 2 (37433_at), score: -0.31 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.4 PLCL2phospholipase C-like 2 (213309_at), score: -0.53 PLGLB2plasminogen-like B2 (205871_at), score: -0.31 POGZpogo transposable element with ZNF domain (215281_x_at), score: -0.35 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: -0.3 POM121L9PPOM121 membrane glycoprotein-like 9 (rat) pseudogene (222253_s_at), score: -0.32 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: -0.36 PPLperiplakin (203407_at), score: -0.51 PRDM2PR domain containing 2, with ZNF domain (203057_s_at), score: -0.3 PRINSpsoriasis associated RNA induced by stress (non-protein coding) (216051_x_at), score: -0.44 PROS1protein S (alpha) (207808_s_at), score: -0.35 PRPF39PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae) (220553_s_at), score: -0.32 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.43 PTGESprostaglandin E synthase (210367_s_at), score: -0.66 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.37 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: -0.39 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: -0.61 RAB11FIP1RAB11 family interacting protein 1 (class I) (219681_s_at), score: -0.49 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.29 RAPGEF2Rap guanine nucleotide exchange factor (GEF) 2 (203097_s_at), score: -0.54 RBM12RNA binding motif protein 12 (212170_at), score: -0.33 RCL1RNA terminal phosphate cyclase-like 1 (218544_s_at), score: -0.42 RGL1ral guanine nucleotide dissociation stimulator-like 1 (209568_s_at), score: -0.4 RGS2regulator of G-protein signaling 2, 24kDa (202388_at), score: -0.29 RGS5regulator of G-protein signaling 5 (218353_at), score: -0.65 RIMS2regulating synaptic membrane exocytosis 2 (215478_at), score: -0.33 RIN2Ras and Rab interactor 2 (209684_at), score: -0.34 RIPK2receptor-interacting serine-threonine kinase 2 (209545_s_at), score: -0.47 RNF139ring finger protein 139 (209510_at), score: -0.32 RNF38ring finger protein 38 (218528_s_at), score: -0.4 RNMTRNA (guanine-7-) methyltransferase (202683_s_at), score: -0.32 ROBO1roundabout, axon guidance receptor, homolog 1 (Drosophila) (213194_at), score: -0.29 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.28 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.3 RPL21P37ribosomal protein L21 pseudogene 37 (216479_at), score: -0.43 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: -0.48 RPL35Aribosomal protein L35a (215208_x_at), score: -0.41 RPLP2ribosomal protein, large, P2 (200908_s_at), score: -0.37 RPS10P3ribosomal protein S10 pseudogene 3 (217336_at), score: -0.33 RPS19ribosomal protein S19 (202648_at), score: -0.38 RPS20ribosomal protein S20 (216247_at), score: -0.39 RPS3AP47ribosomal protein S3a pseudogene 47 (216823_at), score: -0.34 RRADRas-related associated with diabetes (204802_at), score: -0.43 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (214764_at), score: -0.54 RUNX1runt-related transcription factor 1 (209360_s_at), score: -0.6 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.38 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: -0.46 SCD5stearoyl-CoA desaturase 5 (220232_at), score: -0.36 SCML2sex comb on midleg-like 2 (Drosophila) (206147_x_at), score: -0.32 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.34 SERF1Bsmall EDRK-rich factor 1B (centromeric) (219982_s_at), score: -0.42 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.34 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: -0.3 SFRS11splicing factor, arginine/serine-rich 11 (213742_at), score: -0.43 SFRS18splicing factor, arginine/serine-rich 18 (212176_at), score: -0.42 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: -0.3 SH3BP2SH3-domain binding protein 2 (217257_at), score: -0.65 SH3GL3SH3-domain GRB2-like 3 (211565_at), score: -0.42 SHOX2short stature homeobox 2 (210135_s_at), score: -0.32 SIAH1seven in absentia homolog 1 (Drosophila) (202981_x_at), score: -0.32 SKP1S-phase kinase-associated protein 1 (200719_at), score: -0.37 SLC17A6solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (220551_at), score: -0.41 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (209681_at), score: -0.67 SLC2A14solute carrier family 2 (facilitated glucose transporter), member 14 (222088_s_at), score: -0.32 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: -0.31 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: -0.4 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: -0.45 SLC35F2solute carrier family 35, member F2 (218826_at), score: -0.39 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.46 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (205921_s_at), score: -0.46 SLC7A7solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 (204588_s_at), score: -0.36 SLKSTE20-like kinase (yeast) (206875_s_at), score: -0.33 SMEK2SMEK homolog 2, suppressor of mek1 (Dictyostelium) (222270_at), score: -0.45 SMOXspermine oxidase (210357_s_at), score: -0.68 SNIP1Smad nuclear interacting protein 1 (219409_at), score: -0.37 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: -0.85 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: -0.33 SOX4SRY (sex determining region Y)-box 4 (201417_at), score: -0.52 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: -0.49 SPNsialophorin (206057_x_at), score: -0.57 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: -0.83 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: -0.3 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: -0.65 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.32 SPSB1splA/ryanodine receptor domain and SOCS box containing 1 (219677_at), score: -0.34 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: -0.53 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: -0.34 STC1stanniocalcin 1 (204597_x_at), score: -0.52 STK38Lserine/threonine kinase 38 like (212572_at), score: -0.67 SVILsupervillin (202565_s_at), score: -0.56 SYCP1synaptonemal complex protein 1 (206740_x_at), score: -0.56 TAAR2trace amine associated receptor 2 (221394_at), score: -0.42 TAF5LTAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa (213654_at), score: -0.33 TBC1D12TBC1 domain family, member 12 (221858_at), score: -0.29 TBX3T-box 3 (219682_s_at), score: -0.62 TFAP2Ctranscription factor AP-2 gamma (activating enhancer binding protein 2 gamma) (205286_at), score: -0.43 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: -0.3 THBDthrombomodulin (203887_s_at), score: -0.64 THBS2thrombospondin 2 (203083_at), score: -0.39 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: -0.5 TMEM38Btransmembrane protein 38B (218772_x_at), score: -0.38 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: -0.33 TNFAIP3tumor necrosis factor, alpha-induced protein 3 (202644_s_at), score: -0.47 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: -0.43 TP53TG3TP53 target 3 (220167_s_at), score: -0.5 TRIM36tripartite motif-containing 36 (219736_at), score: -0.65 TRIM8tripartite motif-containing 8 (221012_s_at), score: -0.36 TRIT1tRNA isopentenyltransferase 1 (218617_at), score: -0.29 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: -0.48 TULP4tubby like protein 4 (218184_at), score: -0.46 UBE2D1ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) (211764_s_at), score: -0.43 UBQLN4ubiquilin 4 (222252_x_at), score: -0.33 UGT2B28UDP glucuronosyltransferase 2 family, polypeptide B28 (211682_x_at), score: -0.52 UPP1uridine phosphorylase 1 (203234_at), score: -0.46 USP12ubiquitin specific peptidase 12 (213327_s_at), score: -0.67 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: -0.56 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: -0.44 VEZTvezatin, adherens junctions transmembrane protein (207263_x_at), score: -0.29 WASF3WAS protein family, member 3 (204042_at), score: -0.41 WDR43WD repeat domain 43 (214662_at), score: -0.41 WNT6wingless-type MMTV integration site family, member 6 (71933_at), score: -0.37 WRNWerner syndrome (205667_at), score: -0.31 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: -0.29 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: -0.73 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: -0.41 ZCCHC14zinc finger, CCHC domain containing 14 (212655_at), score: -0.32 ZCCHC2zinc finger, CCHC domain containing 2 (219062_s_at), score: -0.59 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: -0.35 ZEB2zinc finger E-box binding homeobox 2 (203603_s_at), score: -0.4 ZFC3H1zinc finger, C3H1-type containing (213065_at), score: -0.42 ZFP106zinc finger protein 106 homolog (mouse) (217781_s_at), score: -0.32 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.47 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: -0.39 ZNF365zinc finger protein 365 (206448_at), score: -0.35 ZNF432zinc finger protein 432 (219848_s_at), score: -0.44 ZNF468zinc finger protein 468 (214751_at), score: -0.44 ZNF492zinc finger protein 492 (215532_x_at), score: -0.37 ZNF493zinc finger protein 493 (211064_at), score: -0.46 ZNF506zinc finger protein 506 (221626_at), score: -0.34 ZNF528zinc finger protein 528 (215019_x_at), score: -0.36 ZNF639zinc finger protein 639 (218413_s_at), score: -0.37 ZNF75Dzinc finger protein 75D (213659_at), score: -0.3 ZNF770zinc finger protein 770 (220608_s_at), score: -0.59 ZNF816Azinc finger protein 816A (217541_x_at), score: -0.5 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: -0.29
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485731.cel | 5 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |