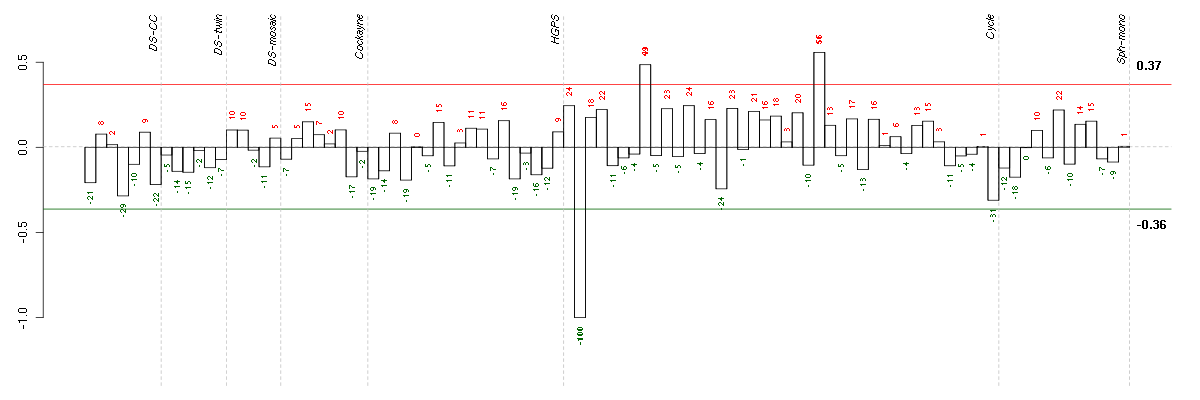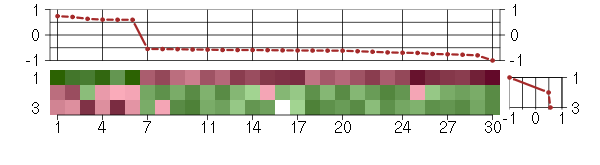

Under-expression is coded with green,
over-expression with red color.



ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -1 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.66 ADRA1Dadrenergic, alpha-1D-, receptor (210961_s_at), score: -0.56 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.8 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.61 CATSPER2cation channel, sperm associated 2 (217588_at), score: -0.61 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.55 CYorf15Bchromosome Y open reading frame 15B (214131_at), score: -0.64 EPS8L1EPS8-like 1 (218778_x_at), score: -0.59 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.76 FXYD2FXYD domain containing ion transport regulator 2 (207434_s_at), score: -0.55 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (203178_at), score: 0.71 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.6 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.74 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.59 ITPR1inositol 1,4,5-triphosphate receptor, type 1 (203710_at), score: 0.6 KCTD17potassium channel tetramerisation domain containing 17 (205561_at), score: -0.59 KIF16Bkinesin family member 16B (219570_at), score: 0.6 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -0.7 LOC100128919similar to HSPC157 (219865_at), score: 0.74 LOC100133748similar to GTF2IRD2 protein (215569_at), score: -0.62 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.61 PHF2PHD finger protein 2 (212726_at), score: 0.6 RBM38RNA binding motif protein 38 (212430_at), score: -0.57 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: -0.7 SCLYselenocysteine lyase (221575_at), score: 0.63 SIX6SIX homeobox 6 (207250_at), score: -0.68 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.57 SYT5synaptotagmin V (206161_s_at), score: -0.59 TMPRSS6transmembrane protease, serine 6 (214955_at), score: -0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |