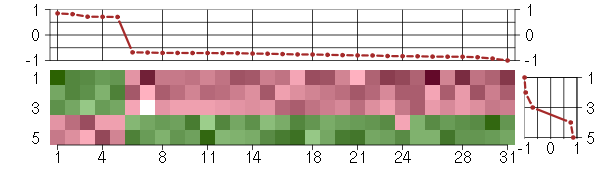

Under-expression is coded with green,
over-expression with red color.



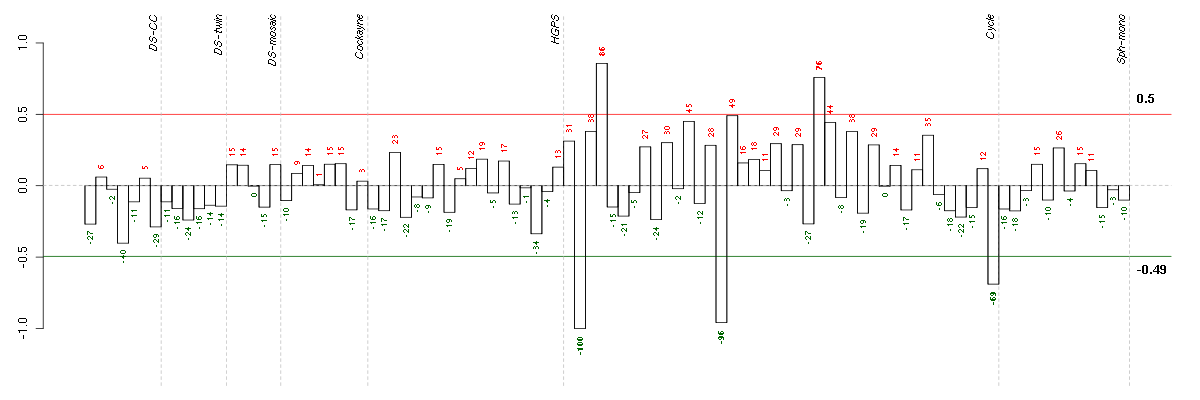
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.85 ADORA1adenosine A1 receptor (216220_s_at), score: -0.7 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.69 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: -0.71 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: 0.72 CCDC40coiled-coil domain containing 40 (220592_at), score: -0.83 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.83 CNNM3cyclin M3 (220739_s_at), score: 0.71 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.68 EPS8L1EPS8-like 1 (218778_x_at), score: -0.76 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -1 HAB1B1 for mucin (215778_x_at), score: -0.79 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: -0.85 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: -0.85 LILRA5leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 (215838_at), score: -0.87 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.73 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: -0.71 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.75 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.73 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.71 PHF2PHD finger protein 2 (212726_at), score: 0.85 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.7 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.92 PRLHprolactin releasing hormone (221443_x_at), score: -0.76 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.71 RBM12BRNA binding motif protein 12B (51228_at), score: 0.82 RBM38RNA binding motif protein 38 (212430_at), score: -0.78 RNF121ring finger protein 121 (219021_at), score: -0.71 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.8 TMPRSS6transmembrane protease, serine 6 (214955_at), score: -0.83 TULP2tubby like protein 2 (206733_at), score: -0.8
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486111.cel | 24 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |