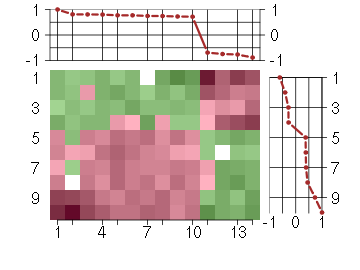

Under-expression is coded with green,
over-expression with red color.



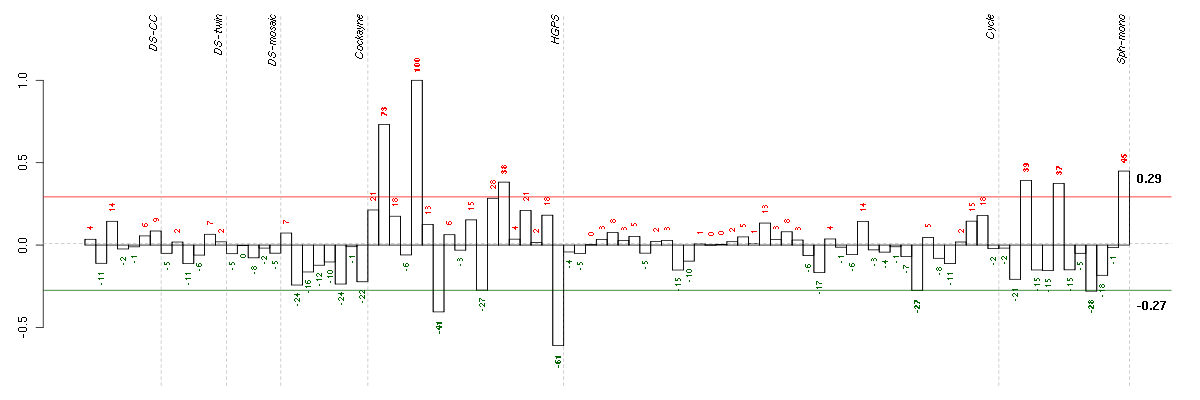
AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.88 CHN1chimerin (chimaerin) 1 (212624_s_at), score: 0.74 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.77 FGL2fibrinogen-like 2 (204834_at), score: -0.69 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.76 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.77 KRT34keratin 34 (206969_at), score: 0.72 KRTAP1-1keratin associated protein 1-1 (220976_s_at), score: 1 OXTRoxytocin receptor (206825_at), score: 0.73 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.75 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: -0.75 STMN2stathmin-like 2 (203000_at), score: 0.8 TNFRSF6Btumor necrosis factor receptor superfamily, member 6b, decoy (206467_x_at), score: 0.81 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 0.81
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |