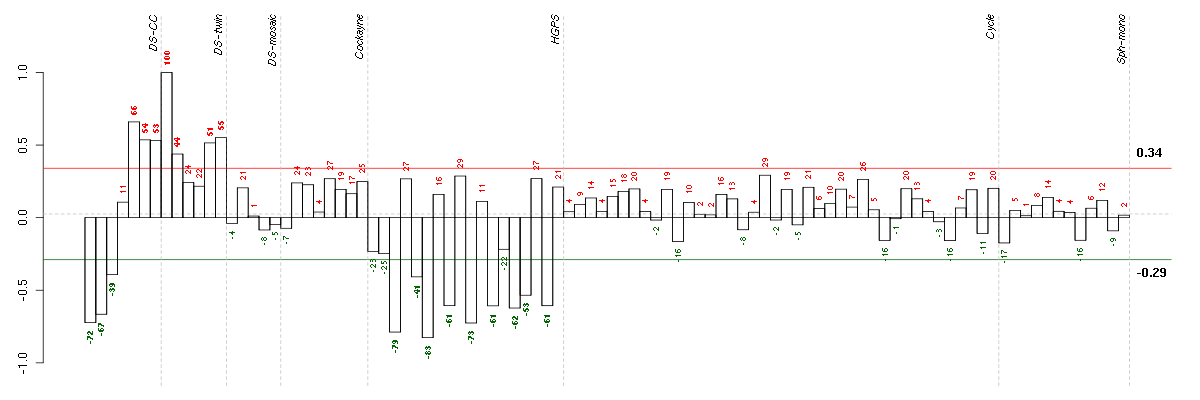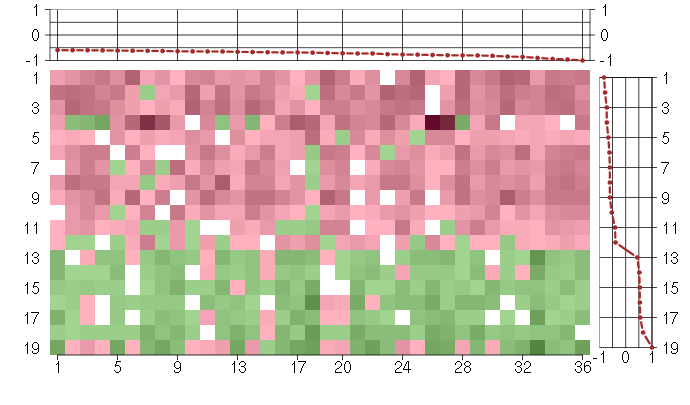

Under-expression is coded with green,
over-expression with red color.



ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: -0.8 AUTS2autism susceptibility candidate 2 (212599_at), score: -0.68 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: -0.8 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: -0.72 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.64 CLGNcalmegin (205830_at), score: -0.64 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: -0.78 DNM1dynamin 1 (215116_s_at), score: -0.65 DPTdermatopontin (213068_at), score: -0.62 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: -0.8 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.61 FHOD3formin homology 2 domain containing 3 (218980_at), score: -0.6 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.6 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: -0.9 HMOX1heme oxygenase (decycling) 1 (203665_at), score: -1 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: -0.6 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: -0.66 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: -0.69 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.72 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: -0.77 LRRC15leucine rich repeat containing 15 (213909_at), score: -0.96 MSX1msh homeobox 1 (205932_s_at), score: -0.82 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.67 NET1neuroepithelial cell transforming 1 (201830_s_at), score: -0.86 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: -0.63 NKX3-1NK3 homeobox 1 (209706_at), score: -0.85 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: -0.68 PSG4pregnancy specific beta-1-glycoprotein 4 (208191_x_at), score: -0.75 RGS3regulator of G-protein signaling 3 (203823_at), score: -0.68 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: -0.71 SLCO3A1solute carrier organic anion transporter family, member 3A1 (219229_at), score: -0.65 STMN2stathmin-like 2 (203000_at), score: -0.77 TBX3T-box 3 (219682_s_at), score: -0.72 TGFAtransforming growth factor, alpha (205016_at), score: -0.62 TMEM158transmembrane protein 158 (213338_at), score: -0.94 TMEM41Btransmembrane protein 41B (212623_at), score: -0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |