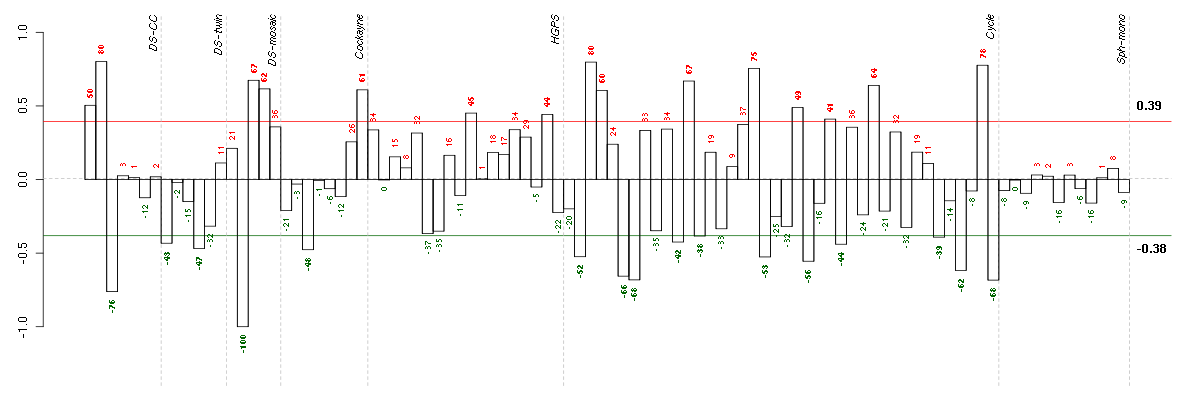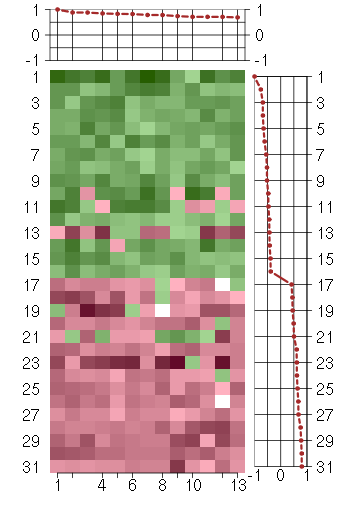

Under-expression is coded with green,
over-expression with red color.



ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.79 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.71 ATF5activating transcription factor 5 (204999_s_at), score: 0.74 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.69 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.84 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.88 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.71 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.82 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.78 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.83 PXNpaxillin (211823_s_at), score: 1 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.88 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.71
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |