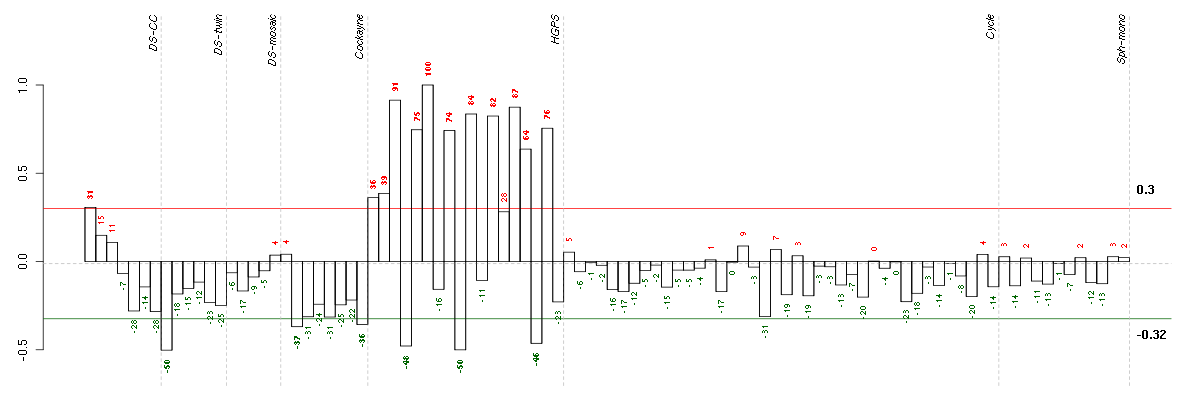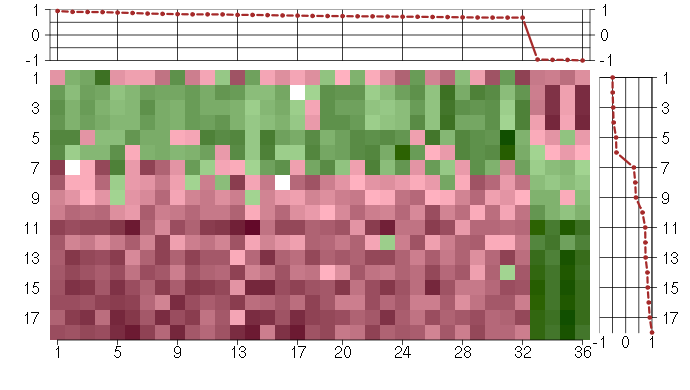

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible


AUTS2autism susceptibility candidate 2 (212599_at), score: 0.81 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.74 C5orf23chromosome 5 open reading frame 23 (219054_at), score: 0.69 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: 0.88 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.68 COMPcartilage oligomeric matrix protein (205713_s_at), score: 0.79 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: 0.77 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: 0.72 FHOD3formin homology 2 domain containing 3 (218980_at), score: 0.73 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.78 GPR116G protein-coupled receptor 116 (212950_at), score: -1 HAS2hyaluronan synthase 2 (206432_at), score: 0.68 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.9 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: 0.81 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.94 KCNN4potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 (204401_at), score: 0.76 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.91 MEOX2mesenchyme homeobox 2 (206201_s_at), score: -0.98 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: 0.7 MSX1msh homeobox 1 (205932_s_at), score: 0.84 MT1Mmetallothionein 1M (217546_at), score: 0.75 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.71 NDNnecdin homolog (mouse) (209550_at), score: 0.68 NET1neuroepithelial cell transforming 1 (201830_s_at), score: 0.82 NKX3-1NK3 homeobox 1 (209706_at), score: 0.79 NRG1neuregulin 1 (206343_s_at), score: 0.72 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: 0.73 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.69 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: -0.97 SFRP1secreted frizzled-related protein 1 (202036_s_at), score: 0.74 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.81 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: 0.86 STMN2stathmin-like 2 (203000_at), score: 0.83 TMEM158transmembrane protein 158 (213338_at), score: 0.9 ZNF423zinc finger protein 423 (214761_at), score: -0.96 ZNF536zinc finger protein 536 (206403_at), score: 0.72
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690215.cel | 2 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |