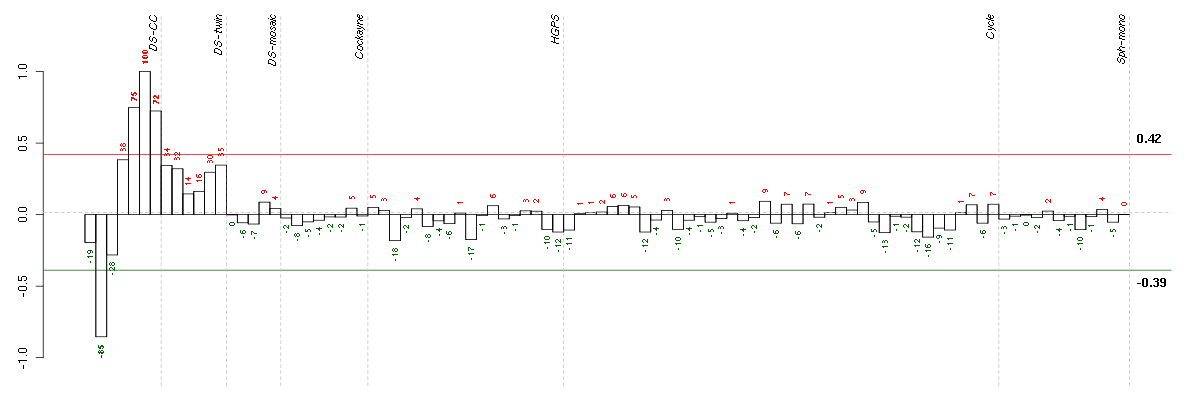

Under-expression is coded with green,
over-expression with red color.

cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: -0.65 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: -0.5 APODapolipoprotein D (201525_at), score: -0.74 ASAP2ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (206414_s_at), score: -0.62 ATF5activating transcription factor 5 (204999_s_at), score: -0.45 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (206233_at), score: 0.49 CADM1cell adhesion molecule 1 (209031_at), score: 0.48 CCND2cyclin D2 (200953_s_at), score: 0.71 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: -0.66 CDH18cadherin 18, type 2 (206280_at), score: 0.55 COL11A1collagen, type XI, alpha 1 (37892_at), score: 0.5 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.58 CSF1colony stimulating factor 1 (macrophage) (209716_at), score: -0.49 CSRP2cysteine and glycine-rich protein 2 (211126_s_at), score: 0.5 CTSBcathepsin B (213274_s_at), score: -0.5 CTSL1cathepsin L1 (202087_s_at), score: -0.54 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: -0.67 EDIL3EGF-like repeats and discoidin I-like domains 3 (207379_at), score: 0.5 EGFRepidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) (201983_s_at), score: -0.47 EIF5Aeukaryotic translation initiation factor 5A (201123_s_at), score: 0.53 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: -0.44 FLGfilaggrin (215704_at), score: 0.98 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: 0.69 GOLSYNGolgi-localized protein (218692_at), score: 0.57 HAPLN1hyaluronan and proteoglycan link protein 1 (205523_at), score: 0.56 HK2hexokinase 2 (202934_at), score: -0.46 HOXC10homeobox C10 (218959_at), score: -0.49 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: -1 JAM2junctional adhesion molecule 2 (219213_at), score: 0.78 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: -0.5 KIAA1644KIAA1644 (52837_at), score: -0.54 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (218326_s_at), score: 0.52 LHFPlipoma HMGIC fusion partner (218656_s_at), score: -0.56 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.61 LY96lymphocyte antigen 96 (206584_at), score: -0.45 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: 0.48 MYO1Dmyosin ID (212338_at), score: -0.56 NFIL3nuclear factor, interleukin 3 regulated (203574_at), score: -0.45 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: -0.45 OLFML1olfactomedin-like 1 (217525_at), score: -0.45 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: 0.56 PFKLphosphofructokinase, liver (201102_s_at), score: 0.59 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.45 POSTNperiostin, osteoblast specific factor (210809_s_at), score: 0.53 PPARDperoxisome proliferator-activated receptor delta (37152_at), score: -0.47 PPLperiplakin (203407_at), score: -0.65 RCAN2regulator of calcineurin 2 (203498_at), score: -0.57 RRADRas-related associated with diabetes (204802_at), score: -0.58 SCRG1scrapie responsive protein 1 (205475_at), score: 0.58 SECTM1secreted and transmembrane 1 (213716_s_at), score: -0.48 SFRP4secreted frizzled-related protein 4 (204051_s_at), score: 0.64 SOD3superoxide dismutase 3, extracellular (205236_x_at), score: -0.65 SOX11SRY (sex determining region Y)-box 11 (204914_s_at), score: 0.63 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: -0.51 TNXBtenascin XB (216333_x_at), score: -0.48 WNT2wingless-type MMTV integration site family member 2 (205648_at), score: 0.52
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |