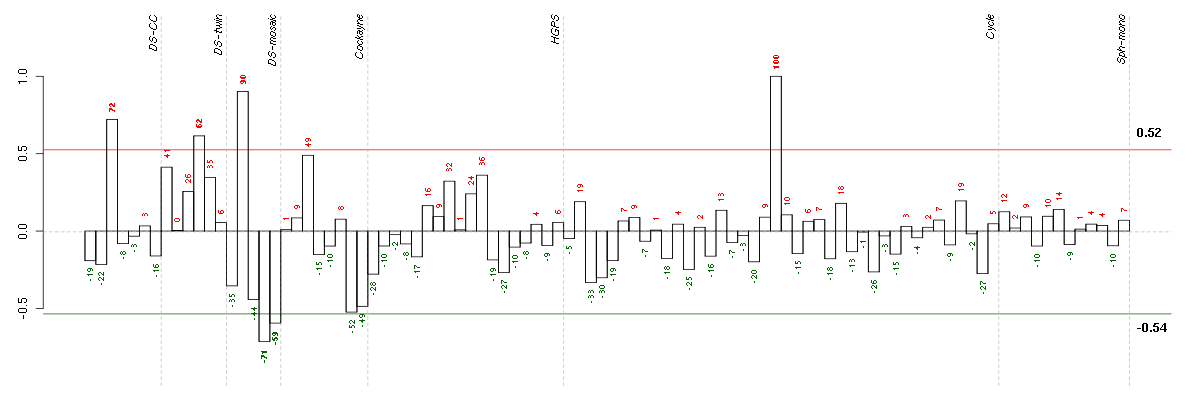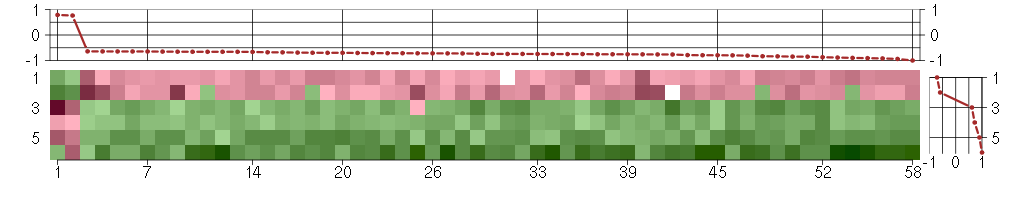

Under-expression is coded with green,
over-expression with red color.



ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: -0.8 ATP13A3ATPase type 13A3 (219558_at), score: -0.71 BCHEbutyrylcholinesterase (205433_at), score: -0.76 BCL2L1BCL2-like 1 (215037_s_at), score: -0.76 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: -0.9 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.74 CDV3CDV3 homolog (mouse) (213548_s_at), score: -0.76 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: 0.76 COL6A1collagen, type VI, alpha 1 (212940_at), score: -0.83 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: -0.91 CPDcarboxypeptidase D (201942_s_at), score: -0.75 CYTH3cytohesin 3 (206523_at), score: -0.64 CYTIPcytohesin 1 interacting protein (209606_at), score: -0.64 DAPK3death-associated protein kinase 3 (203890_s_at), score: -0.68 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: -0.72 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.79 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: -0.88 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: -0.74 FN1fibronectin 1 (214701_s_at), score: -1 GTF2Igeneral transcription factor II, i (210892_s_at), score: -0.74 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.65 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: -0.69 INHBEinhibin, beta E (210587_at), score: 0.79 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: -0.79 LAMP1lysosomal-associated membrane protein 1 (201551_s_at), score: -0.84 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: -0.72 LOXlysyl oxidase (213640_s_at), score: -0.66 LOXL2lysyl oxidase-like 2 (202997_s_at), score: -0.75 MAXMYC associated factor X (210734_x_at), score: -0.85 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: -0.65 MFN2mitofusin 2 (216205_s_at), score: -0.74 MMP14matrix metallopeptidase 14 (membrane-inserted) (202827_s_at), score: -0.72 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: -0.68 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: -0.65 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.92 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: -0.94 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: -0.66 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: -0.7 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: -0.71 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: -0.72 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: -0.66 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: -0.65 PRRX1paired related homeobox 1 (205991_s_at), score: -0.76 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: -0.87 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: -0.76 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: -0.76 PXNpaxillin (211823_s_at), score: -0.85 RAB35RAB35, member RAS oncogene family (205461_at), score: -0.66 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: -0.74 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: -0.7 RXRBretinoid X receptor, beta (215099_s_at), score: -0.72 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: -0.7 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: -0.75 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: -0.66 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: -0.7 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: -0.66 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.81 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: -0.79
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |