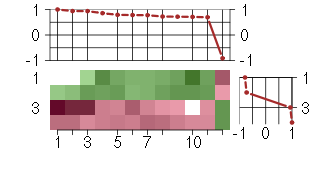

Under-expression is coded with green,
over-expression with red color.



ACTN3actinin, alpha 3 (206891_at), score: 0.71 C2orf54chromosome 2 open reading frame 54 (220149_at), score: 0.86 CALB2calbindin 2 (205428_s_at), score: 0.94 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.78 CYP24A1cytochrome P450, family 24, subfamily A, polypeptide 1 (206504_at), score: 1 EDARectodysplasin A receptor (220048_at), score: 0.72 KRT86keratin 86 (215189_at), score: 0.94 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: -0.91 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.72 NXPH3neurexophilin 3 (221991_at), score: 0.78 OR2W1olfactory receptor, family 2, subfamily W, member 1 (221451_s_at), score: 0.79 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: 0.7
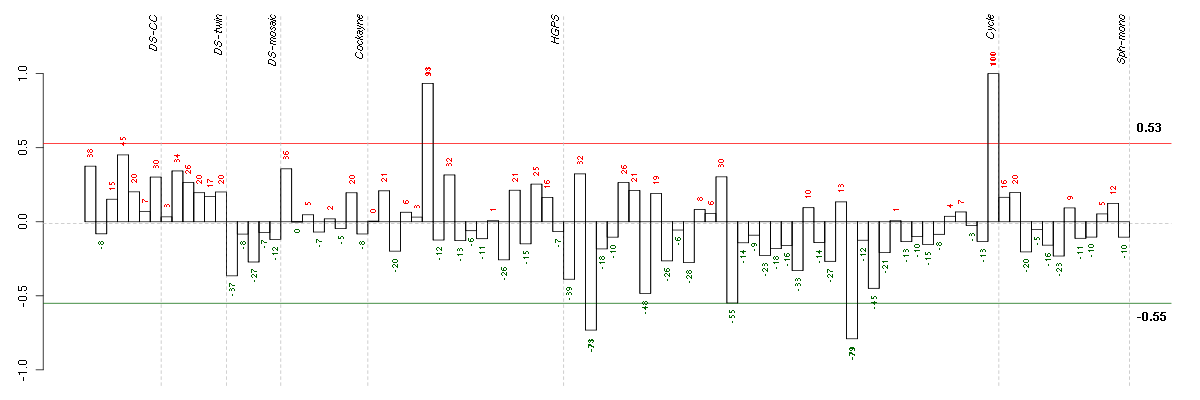
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |