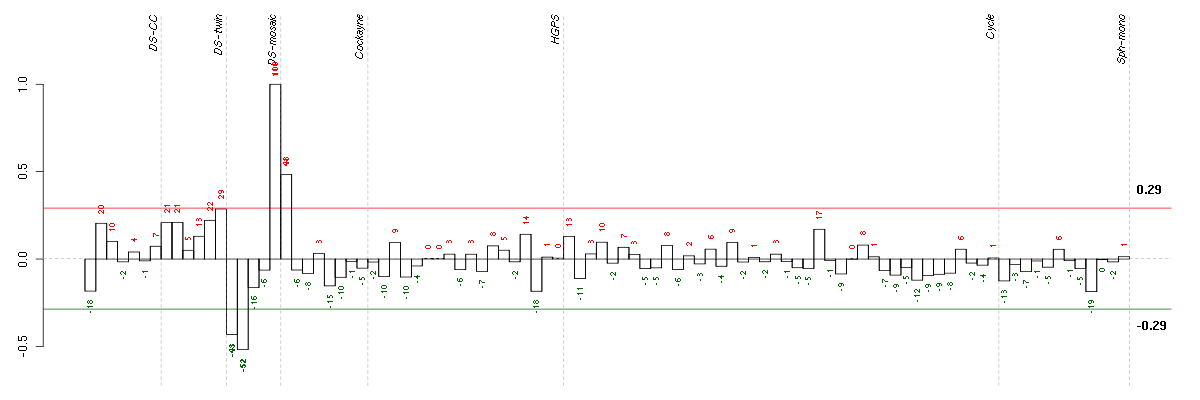

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

APOL1apolipoprotein L, 1 (209546_s_at), score: -0.71 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: -0.72 CA11carbonic anhydrase XI (209726_at), score: -0.84 CFIcomplement factor I (203854_at), score: -0.75 ELOVL2elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 2 (213712_at), score: -0.77 ERAP1endoplasmic reticulum aminopeptidase 1 (214012_at), score: -0.84 FOSv-fos FBJ murine osteosarcoma viral oncogene homolog (209189_at), score: -1 GPC4glypican 4 (204983_s_at), score: -0.78 HHEXhematopoietically expressed homeobox (215933_s_at), score: -0.84 HOXB9homeobox B9 (216417_x_at), score: 0.82 PAPPA2pappalysin 2 (213332_at), score: -0.81 PDE1Cphosphodiesterase 1C, calmodulin-dependent 70kDa (207303_at), score: 0.91 PKP2plakophilin 2 (207717_s_at), score: -0.83 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: -0.83 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: -0.79 SCARB1scavenger receptor class B, member 1 (201819_at), score: -0.84 SCG2secretogranin II (chromogranin C) (204035_at), score: -0.9 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: -0.7 TMEM132Atransmembrane protein 132A (218834_s_at), score: -0.72 ZNF711zinc finger protein 711 (207781_s_at), score: -0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 47C.CEL | 5 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 5 |