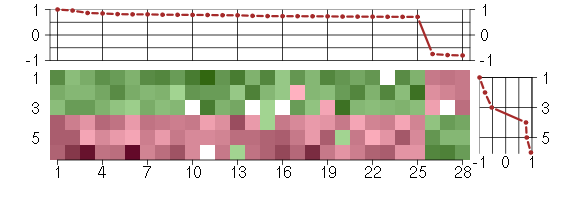

Under-expression is coded with green,
over-expression with red color.



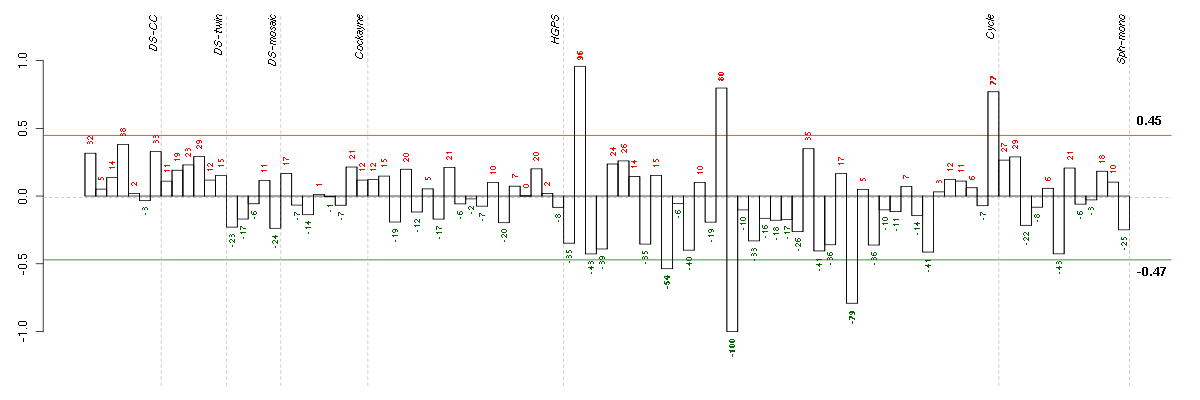
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.81 ACTN3actinin, alpha 3 (206891_at), score: 0.71 ALPPL2alkaline phosphatase, placental-like 2 (216377_x_at), score: 0.79 ATMataxia telangiectasia mutated (210858_x_at), score: 0.82 C19orf40chromosome 19 open reading frame 40 (214816_x_at), score: 0.77 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.79 CCDC40coiled-coil domain containing 40 (220592_at), score: 1 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.74 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.71 F11coagulation factor XI (206610_s_at), score: 0.78 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.96 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: -0.8 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: 0.71 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (211693_at), score: 0.73 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.75 KRT8P12keratin 8 pseudogene 12 (222060_at), score: 0.86 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: 0.75 MYO15Amyosin XVA (220288_at), score: 0.72 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.85 PKLRpyruvate kinase, liver and RBC (222078_at), score: 0.73 RBM38RNA binding motif protein 38 (212430_at), score: 0.79 ROS1c-ros oncogene 1 , receptor tyrosine kinase (207569_at), score: 0.72 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.81 SEPT5septin 5 (209767_s_at), score: 0.78 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.74 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: 0.74 TNFSF10tumor necrosis factor (ligand) superfamily, member 10 (214329_x_at), score: 0.72 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: 0.8
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |