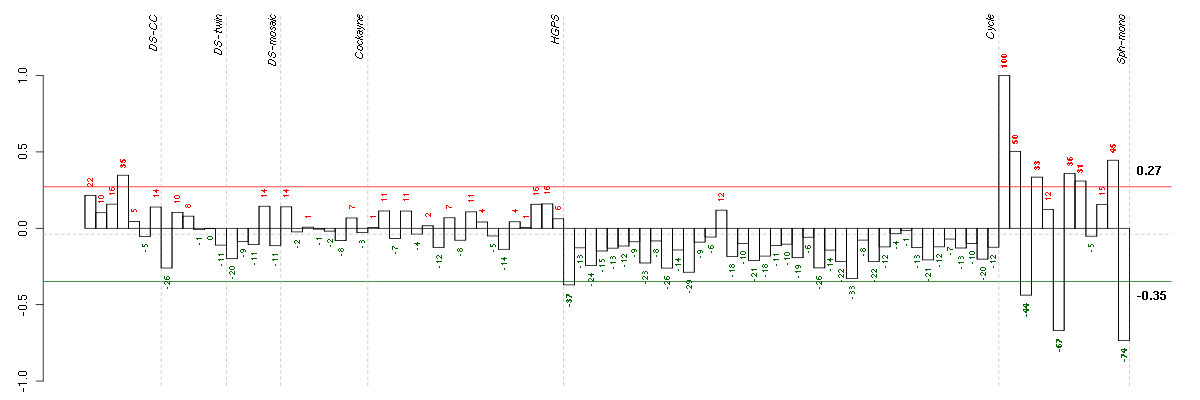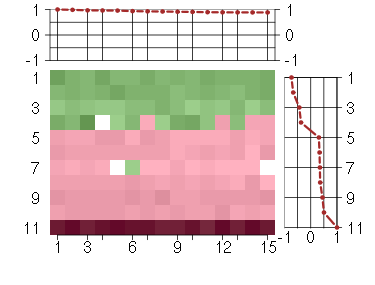

Under-expression is coded with green,
over-expression with red color.



ALMS1Alstrom syndrome 1 (214707_x_at), score: 0.9 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (212972_x_at), score: 0.89 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.97 DNAH3dynein, axonemal, heavy chain 3 (220725_x_at), score: 0.93 FLJ11292hypothetical protein FLJ11292 (220828_s_at), score: 0.96 G3BP1GTPase activating protein (SH3 domain) binding protein 1 (222187_x_at), score: 0.99 METTL7Amethyltransferase like 7A (211424_x_at), score: 0.91 SH3GL3SH3-domain GRB2-like 3 (211565_at), score: 0.88 SLC30A5solute carrier family 30 (zinc transporter), member 5 (220181_x_at), score: 0.94 SPINLW1serine peptidase inhibitor-like, with Kunitz and WAP domains 1 (eppin) (206318_at), score: 0.92 SPNsialophorin (206057_x_at), score: 0.89 SPTLC3serine palmitoyltransferase, long chain base subunit 3 (220456_at), score: 1 TIMM8Atranslocase of inner mitochondrial membrane 8 homolog A (yeast) (210800_at), score: 0.96 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.89 ZNF816Azinc finger protein 816A (217541_x_at), score: 0.89
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |