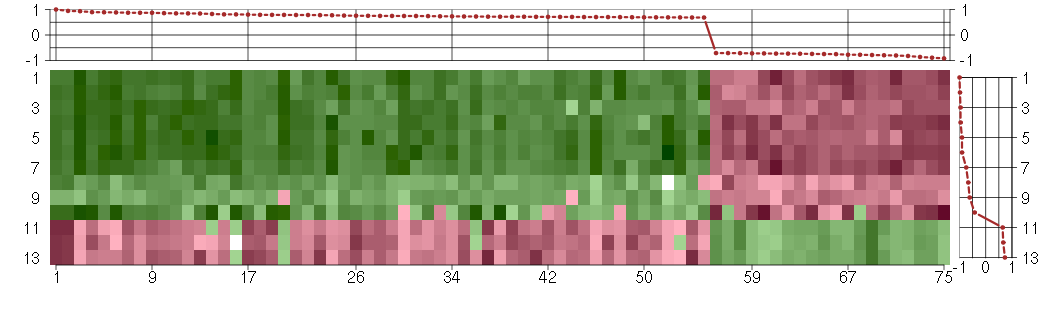

Under-expression is coded with green,
over-expression with red color.

cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
neuron projection
A prolongation or process extending from a nerve cell, e.g. an axon or dendrite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.

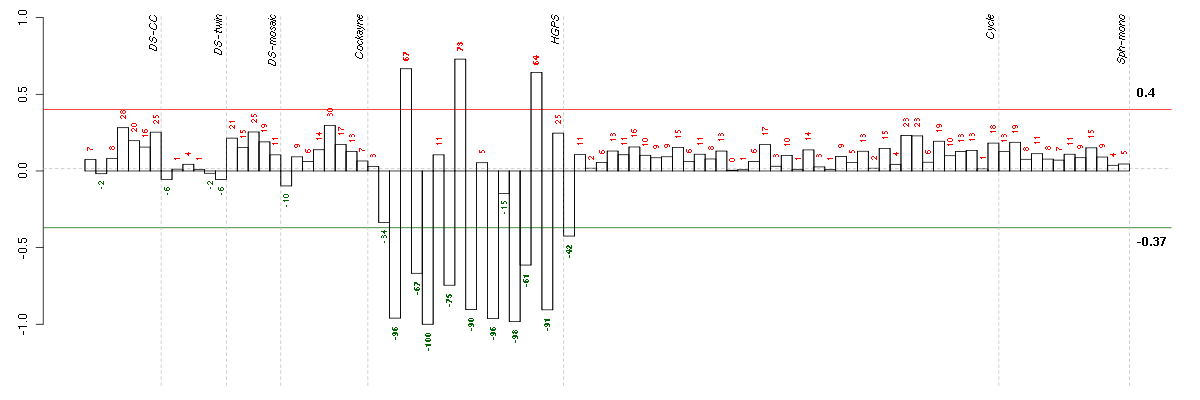
ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: 0.78 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: 0.79 AUTS2autism susceptibility candidate 2 (212599_at), score: -0.79 BST1bone marrow stromal cell antigen 1 (205715_at), score: 0.81 C1orf54chromosome 1 open reading frame 54 (219506_at), score: 0.78 CAMK2N1calcium/calmodulin-dependent protein kinase II inhibitor 1 (218309_at), score: -0.92 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: -0.71 CCDC81coiled-coil domain containing 81 (220389_at), score: 0.71 CCR10chemokine (C-C motif) receptor 10 (220565_at), score: 0.72 CTSCcathepsin C (201487_at), score: 0.7 CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: 0.69 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: -0.75 EPHA5EPH receptor A5 (215664_s_at), score: 0.81 F2Rcoagulation factor II (thrombin) receptor (203989_x_at), score: 0.77 FHOD3formin homology 2 domain containing 3 (218980_at), score: -0.72 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.71 FSTfollistatin (204948_s_at), score: 0.73 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.72 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.7 GPR116G protein-coupled receptor 116 (212950_at), score: 1 HMOX1heme oxygenase (decycling) 1 (203665_at), score: -0.73 HS3ST3A1heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 (219985_at), score: -0.86 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.78 ITGA8integrin, alpha 8 (214265_at), score: 0.72 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: 0.85 KALRNkalirin, RhoGEF kinase (206078_at), score: 0.79 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.75 KLF9Kruppel-like factor 9 (203543_s_at), score: 0.69 KRT18keratin 18 (201596_x_at), score: 0.7 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: 0.74 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (213880_at), score: 0.73 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: 0.84 LPHN2latrophilin 2 (206953_s_at), score: 0.82 LPXNleupaxin (216250_s_at), score: -0.74 LRRC15leucine rich repeat containing 15 (213909_at), score: -0.82 LRRC17leucine rich repeat containing 17 (205381_at), score: 0.73 LRRC32leucine rich repeat containing 32 (203835_at), score: 0.69 MBPmyelin basic protein (210136_at), score: 0.87 MEOX2mesenchyme homeobox 2 (206201_s_at), score: 0.93 MESTmesoderm specific transcript homolog (mouse) (202016_at), score: 0.75 MGPmatrix Gla protein (202291_s_at), score: 0.87 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: -0.79 MSX1msh homeobox 1 (205932_s_at), score: -0.75 NDNnecdin homolog (mouse) (209550_at), score: -0.71 NDUFA4L2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 (218484_at), score: 0.89 NEFMneurofilament, medium polypeptide (205113_at), score: 0.72 NET1neuroepithelial cell transforming 1 (201830_s_at), score: -0.77 NKX3-1NK3 homeobox 1 (209706_at), score: -0.73 NOVnephroblastoma overexpressed gene (214321_at), score: 0.71 NRXN3neurexin 3 (205795_at), score: 0.79 PDE10Aphosphodiesterase 10A (205501_at), score: 0.73 PLA2G16phospholipase A2, group XVI (209581_at), score: 0.69 PLXND1plexin D1 (38671_at), score: 0.69 PNMA2paraneoplastic antigen MA2 (209598_at), score: 0.78 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (218292_s_at), score: 0.75 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: 0.94 RELNreelin (205923_at), score: 0.9 RGS7regulator of G-protein signaling 7 (206290_s_at), score: 0.76 RTN1reticulon 1 (203485_at), score: 0.76 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: -0.74 SLC24A3solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 (57588_at), score: 0.8 SLC25A4solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 (202825_at), score: 0.74 SLC7A8solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (216092_s_at), score: -0.8 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (208608_s_at), score: 0.7 SOBPsine oculis binding protein homolog (Drosophila) (218974_at), score: 0.7 SRGNserglycin (201859_at), score: 0.69 STAT4signal transducer and activator of transcription 4 (206118_at), score: 0.72 STMN2stathmin-like 2 (203000_at), score: -0.71 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: 0.87 TMEM158transmembrane protein 158 (213338_at), score: -0.89 TNXAtenascin XA pseudogene (213451_x_at), score: 0.85 TNXBtenascin XB (216333_x_at), score: 0.86 TRPV2transient receptor potential cation channel, subfamily V, member 2 (219282_s_at), score: 0.71 WWP2WW domain containing E3 ubiquitin protein ligase 2 (204022_at), score: 0.75 ZNF423zinc finger protein 423 (214761_at), score: 0.89
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690392.cel | 14 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690223.cel | 3 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690472.cel | 17 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690336.cel | 9 | 5 | HGPS | hgu133a | HGPS | AG10750 |