
Under-expression is coded with green,
over-expression with red color.

defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
phospholipid binding
Interacting selectively with phospholipids, a class of lipids containing phosphoric acid as a mono- or diester.
calcium-dependent phospholipid binding
Interacting selectively with phospholipids, a class of lipids containing phosphoric acid as a mono- or diester, in the presence of calcium.
lipid binding
Interacting selectively with a lipid.
all
This term is the most general term possible
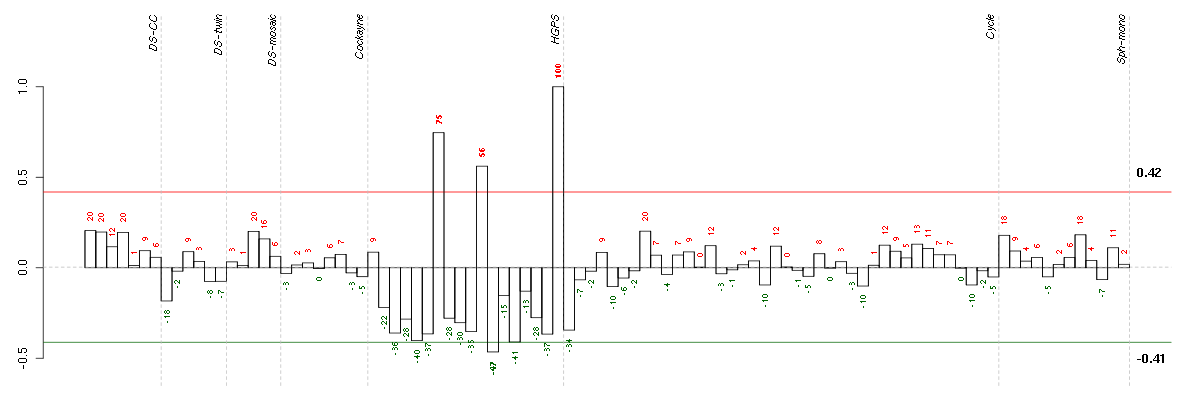
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.73 ADAMTSL3ADAMTS-like 3 (213974_at), score: 0.71 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.61 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (206029_at), score: 0.78 ANXA10annexin A10 (210143_at), score: 0.61 ANXA3annexin A3 (209369_at), score: 0.69 BNC2basonuclin 2 (220272_at), score: 0.64 C8orf84chromosome 8 open reading frame 84 (214725_at), score: 0.82 CD200CD200 molecule (209583_s_at), score: 0.84 CD34CD34 molecule (209543_s_at), score: 0.76 CFHcomplement factor H (213800_at), score: 0.76 CFHR1complement factor H-related 1 (215388_s_at), score: 0.66 CHRDL1chordin-like 1 (209763_at), score: 0.62 CLDN1claudin 1 (218182_s_at), score: 0.66 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: 0.65 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.71 DSG2desmoglein 2 (217901_at), score: 0.62 EPHA5EPH receptor A5 (215664_s_at), score: 0.79 ERGv-ets erythroblastosis virus E26 oncogene homolog (avian) (213541_s_at), score: 0.8 FGL2fibrinogen-like 2 (204834_at), score: 1 FMO3flavin containing monooxygenase 3 (40665_at), score: 0.61 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.61 GPR126G protein-coupled receptor 126 (213094_at), score: 0.7 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.66 GSTT2glutathione S-transferase theta 2 (205439_at), score: 0.7 HYIhydroxypyruvate isomerase homolog (E. coli) (221435_x_at), score: 0.67 IFI27interferon, alpha-inducible protein 27 (202411_at), score: 0.75 IL18interleukin 18 (interferon-gamma-inducing factor) (206295_at), score: 0.69 IL1Ainterleukin 1, alpha (210118_s_at), score: 0.7 ITM2Aintegral membrane protein 2A (202746_at), score: 0.71 LAMC2laminin, gamma 2 (202267_at), score: 0.78 LIPGlipase, endothelial (219181_at), score: 0.9 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: 0.73 MCTP1multiple C2 domains, transmembrane 1 (220122_at), score: 0.64 MYLKmyosin light chain kinase (202555_s_at), score: -0.69 NOX4NADPH oxidase 4 (219773_at), score: 0.73 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: 0.63 PLXNA2plexin A2 (213030_s_at), score: 0.65 POPDC3popeye domain containing 3 (219926_at), score: -0.67 PPM1Hprotein phosphatase 1H (PP2C domain containing) (212686_at), score: 0.68 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: 0.7 SOX17SRY (sex determining region Y)-box 17 (219993_at), score: 0.79 THY1Thy-1 cell surface antigen (208850_s_at), score: -0.66 TLR4toll-like receptor 4 (221060_s_at), score: 0.79 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.7 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: 0.64 ZFHX4zinc finger homeobox 4 (219779_at), score: 0.69 ZNF323zinc finger protein 323 (222016_s_at), score: 0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-3860-raw-cel-1561690480.cel | 18 | 5 | HGPS | hgu133a | HGPS | AG11498 |