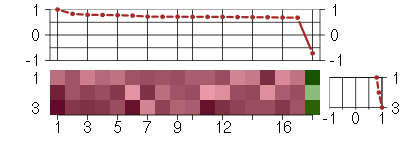

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

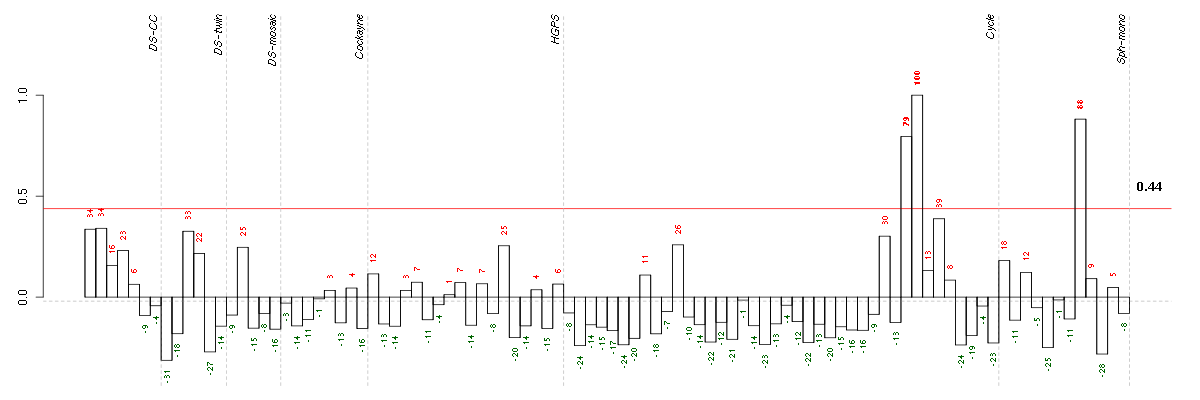
BDKRB2bradykinin receptor B2 (205870_at), score: 0.78 C3complement component 3 (217767_at), score: 0.7 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: 0.72 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.68 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 1 DLX2distal-less homeobox 2 (207147_at), score: -0.72 FMODfibromodulin (202709_at), score: 0.68 GDF15growth differentiation factor 15 (221577_x_at), score: 0.71 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.72 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.83 OMDosteomodulin (205907_s_at), score: 0.69 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.71 PDPNpodoplanin (221898_at), score: 0.7 PPLperiplakin (203407_at), score: 0.76 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: 0.71 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: 0.72 SESN1sestrin 1 (218346_s_at), score: 0.79 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.79
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |