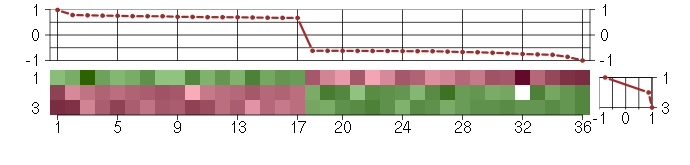

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

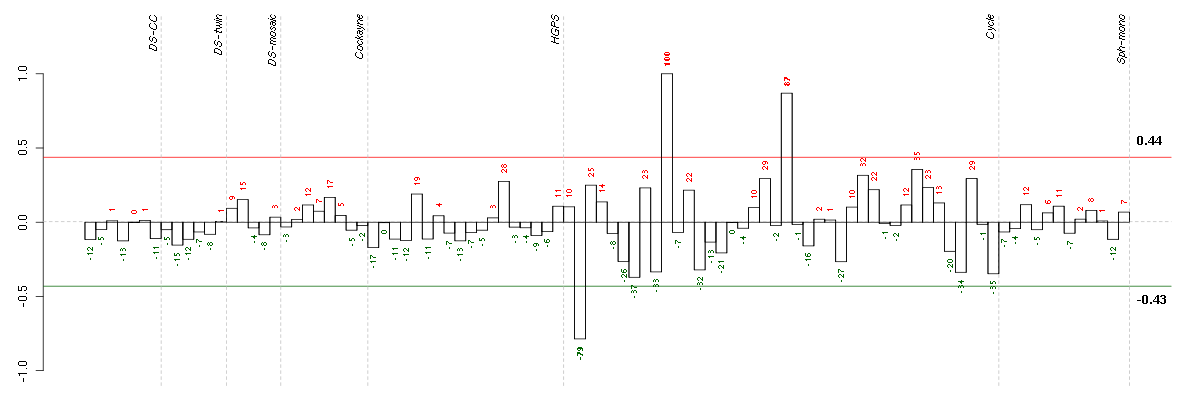
B9D1B9 protein domain 1 (210534_s_at), score: 0.75 CCDC9coiled-coil domain containing 9 (206257_at), score: -0.64 CHST10carbohydrate sulfotransferase 10 (204065_at), score: -0.64 DEPDC6DEP domain containing 6 (218858_at), score: 0.67 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.78 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.86 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -1 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: -0.63 IL33interleukin 33 (209821_at), score: -0.63 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.62 KIAA1305KIAA1305 (220911_s_at), score: 0.71 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: 0.67 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -0.75 LOC646808hypothetical LOC646808 (222338_x_at), score: 0.69 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.68 LTBP4latent transforming growth factor beta binding protein 4 (210628_x_at), score: -0.72 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: -0.78 NDRG4NDRG family member 4 (209159_s_at), score: 0.98 NEFMneurofilament, medium polypeptide (205113_at), score: -0.62 PLAC8placenta-specific 8 (219014_at), score: 0.76 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.65 PRSS3protease, serine, 3 (213421_x_at), score: -0.62 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.74 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.76 SAA4serum amyloid A4, constitutive (207096_at), score: -0.63 SESN1sestrin 1 (218346_s_at), score: 0.7 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.67 SIX6SIX homeobox 6 (207250_at), score: -0.62 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.74 SNX27sorting nexin family member 27 (221006_s_at), score: 0.68 SUOXsulfite oxidase (204067_at), score: 0.71 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.74 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 0.7 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.7 WFDC8WAP four-disulfide core domain 8 (215276_at), score: -0.7 ZNF3zinc finger protein 3 (212684_at), score: 0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |